Below are the slides and materials used to teach the FreeSurfer class during my time at the Stanford Neurodevelopment, Affect, and Psychopathology Lab. It was written for version 5.3, but is compatible with 6.0. Enjoy! Email/message me if you have questions.
GitHub Repository containing all class materials is found here.
FreeSurfer website is found here.


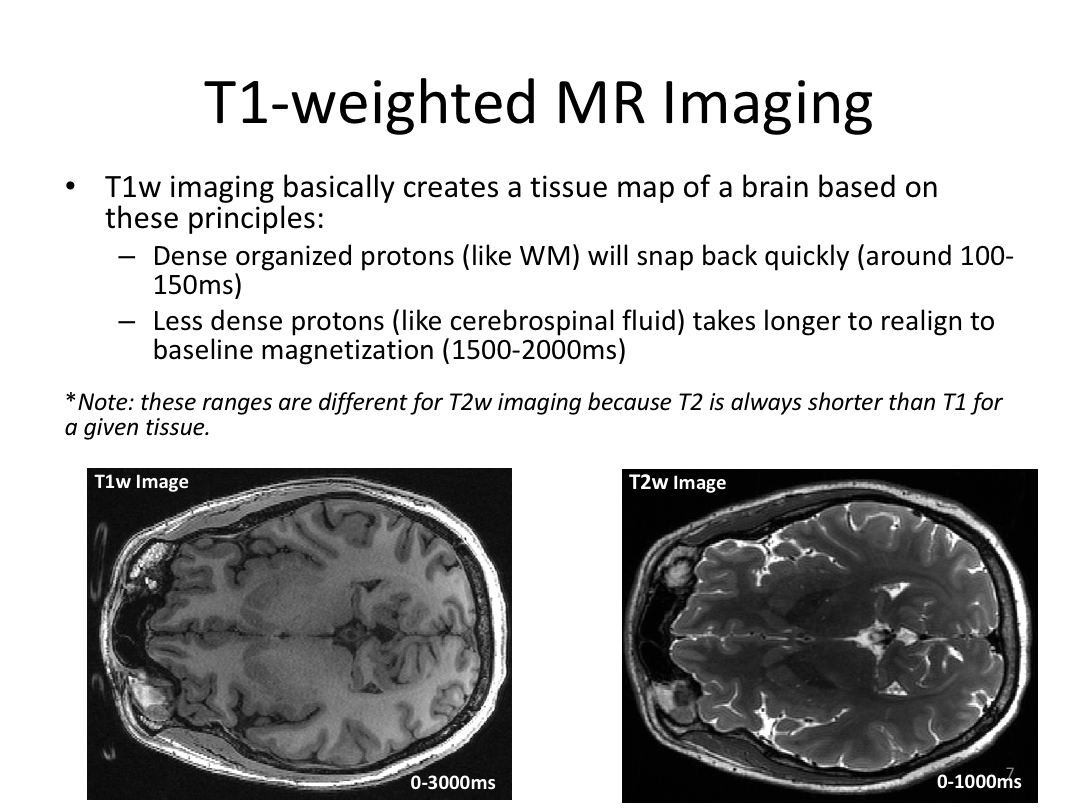
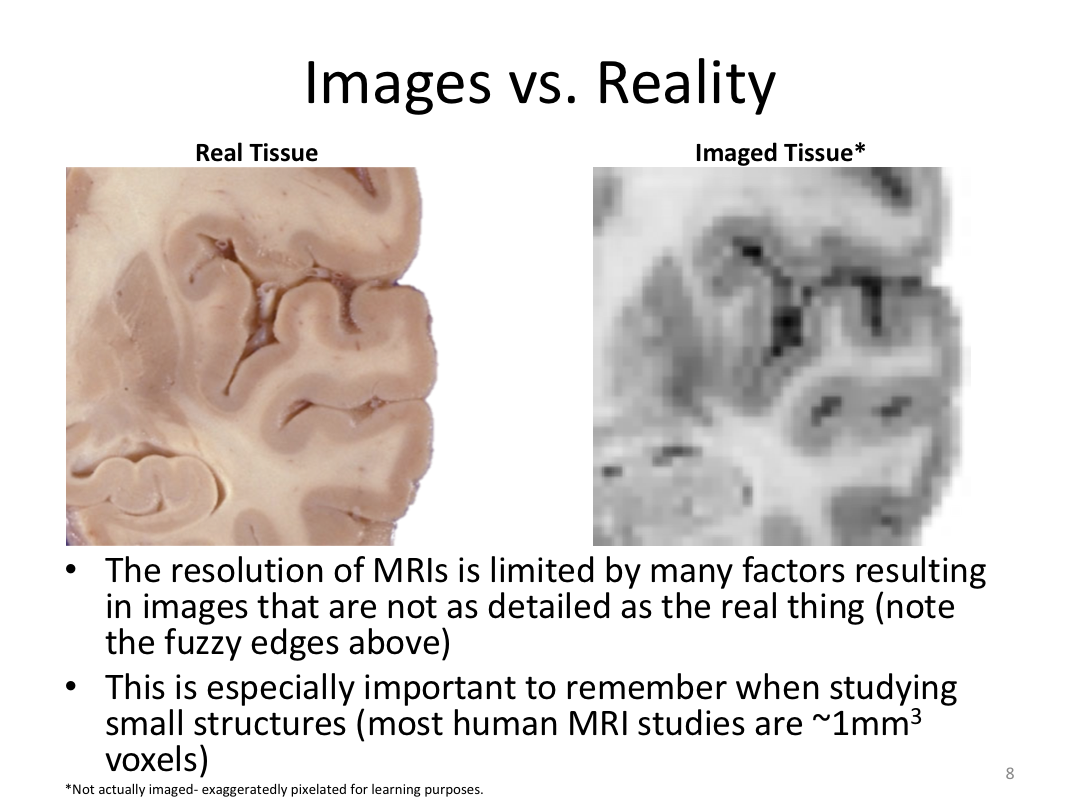
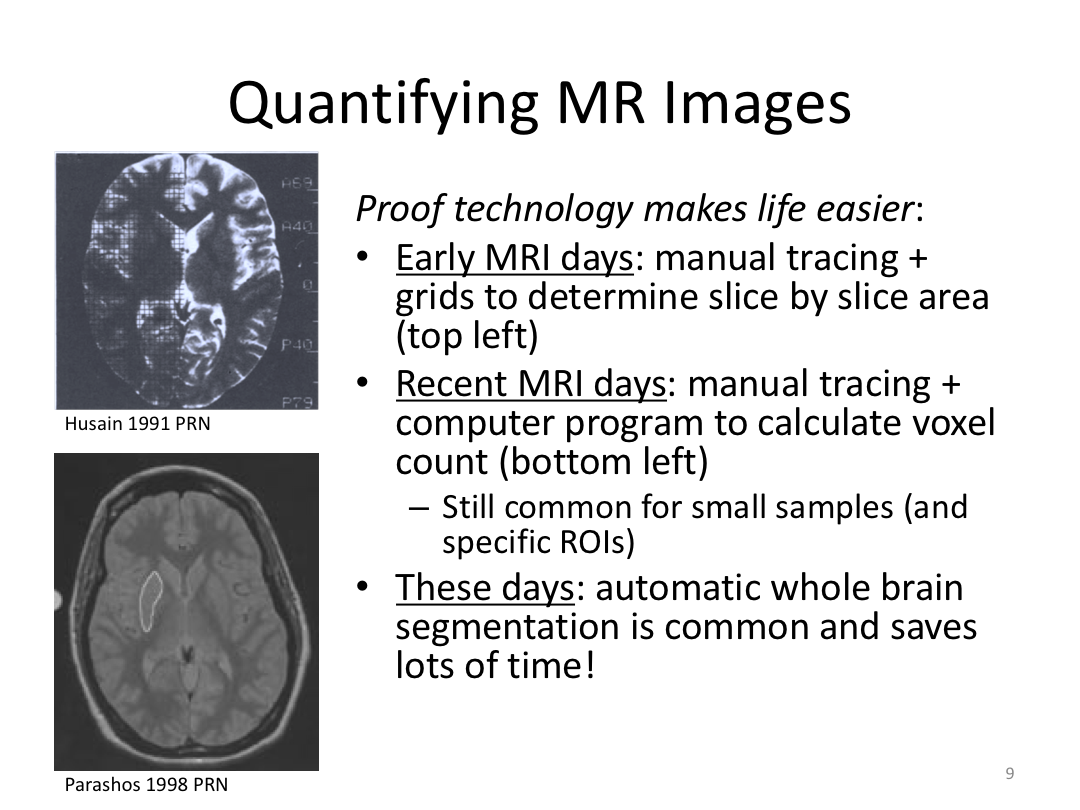
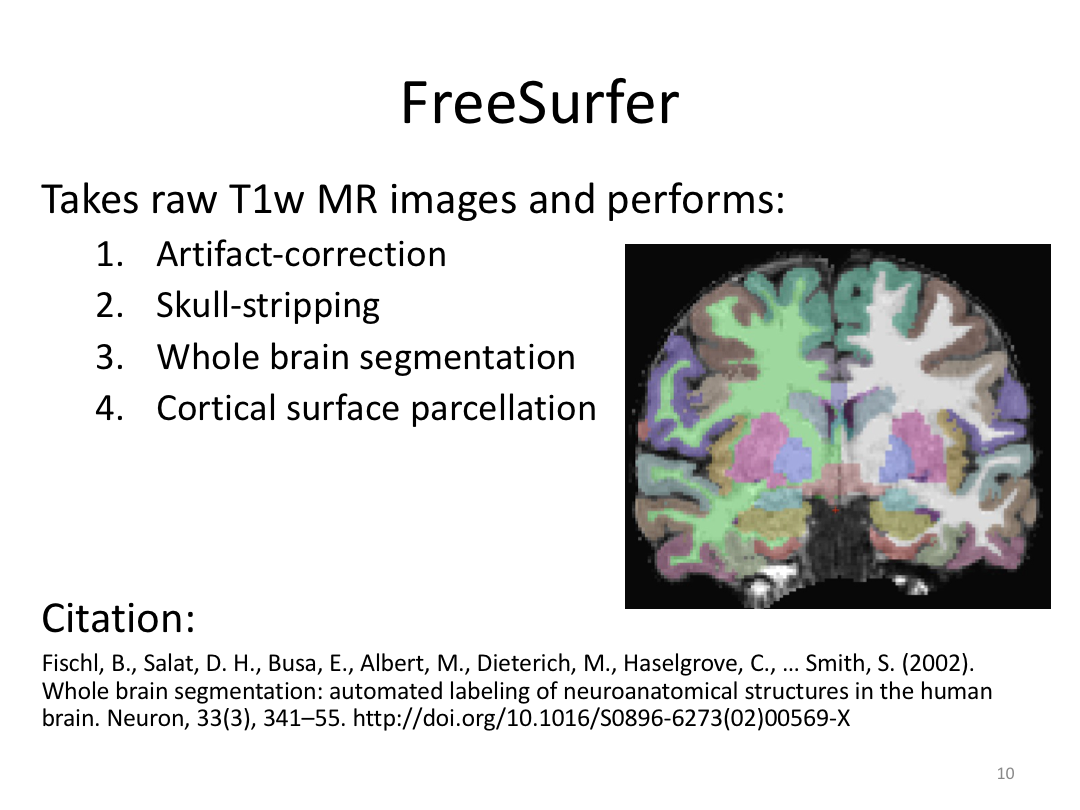


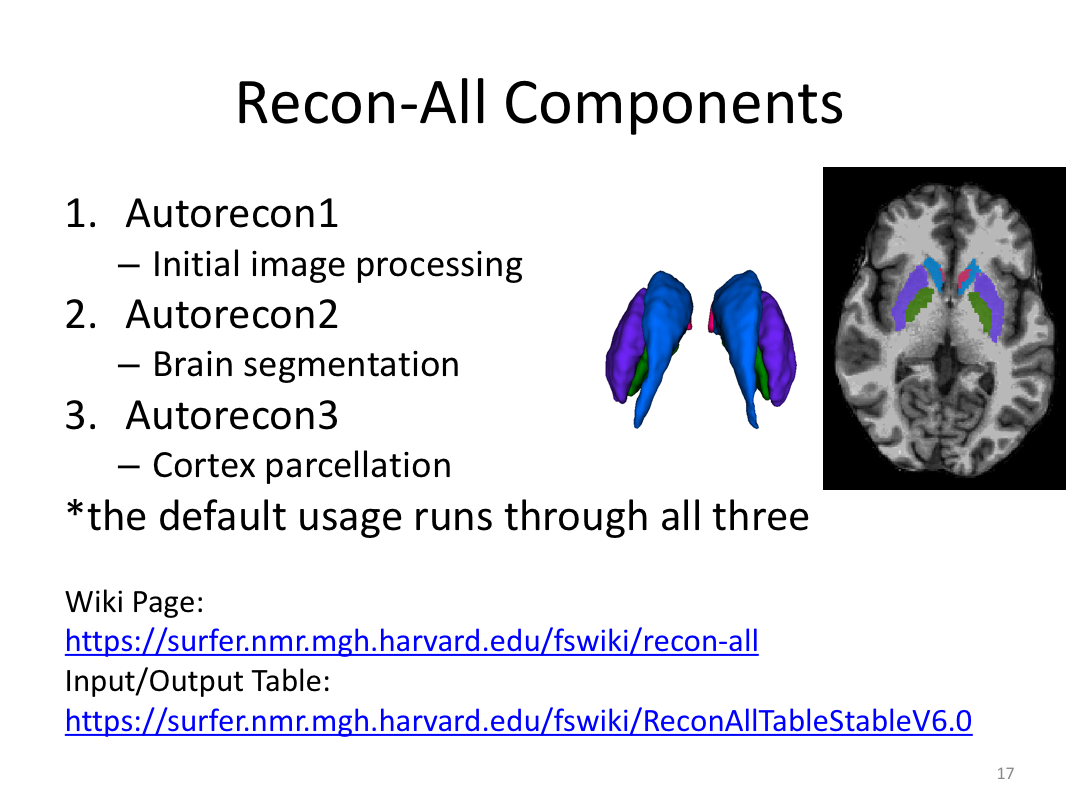
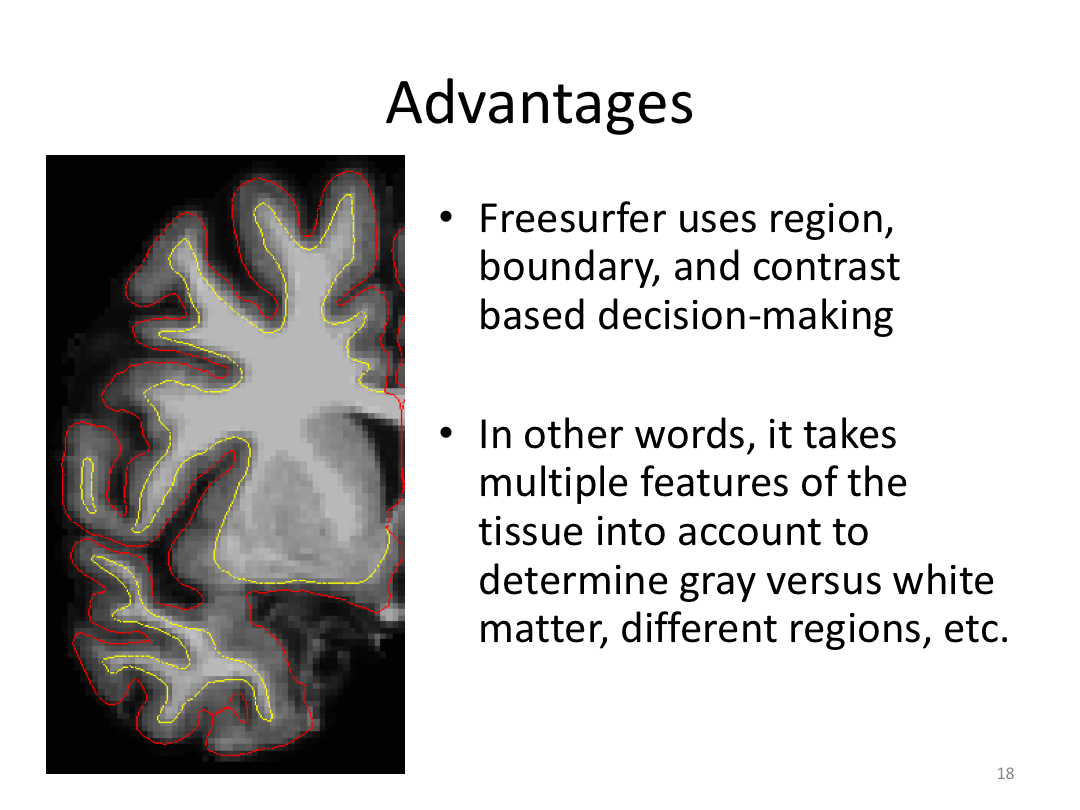

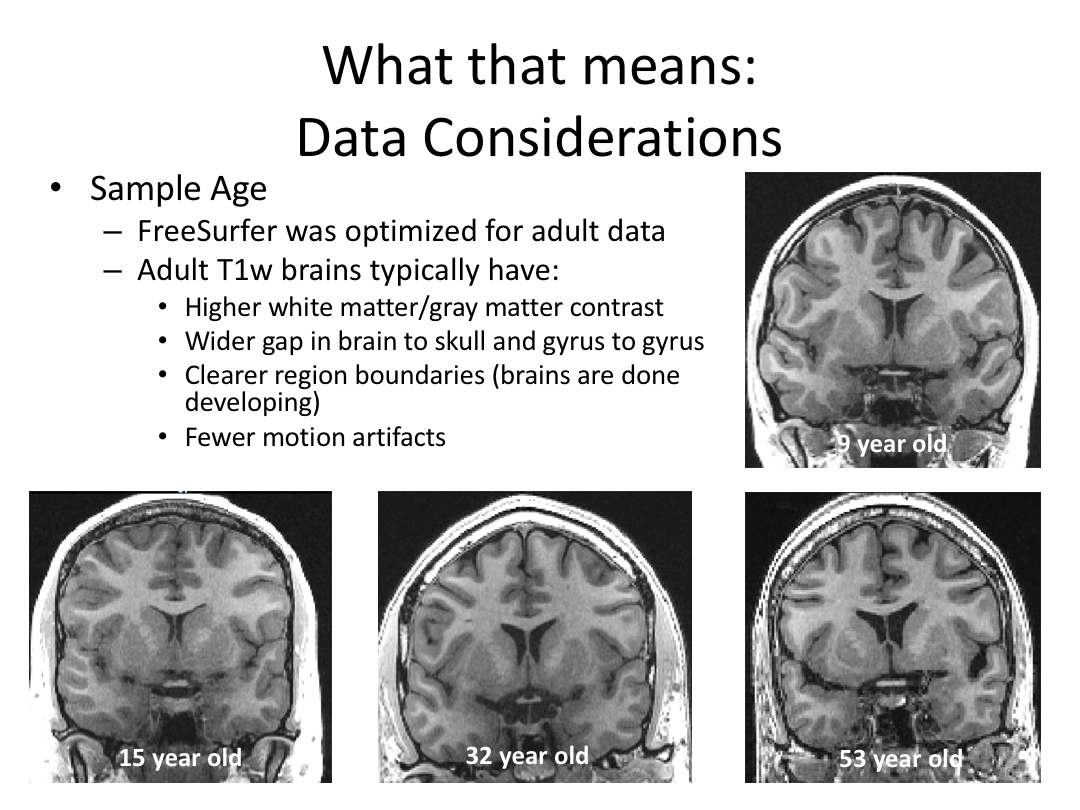
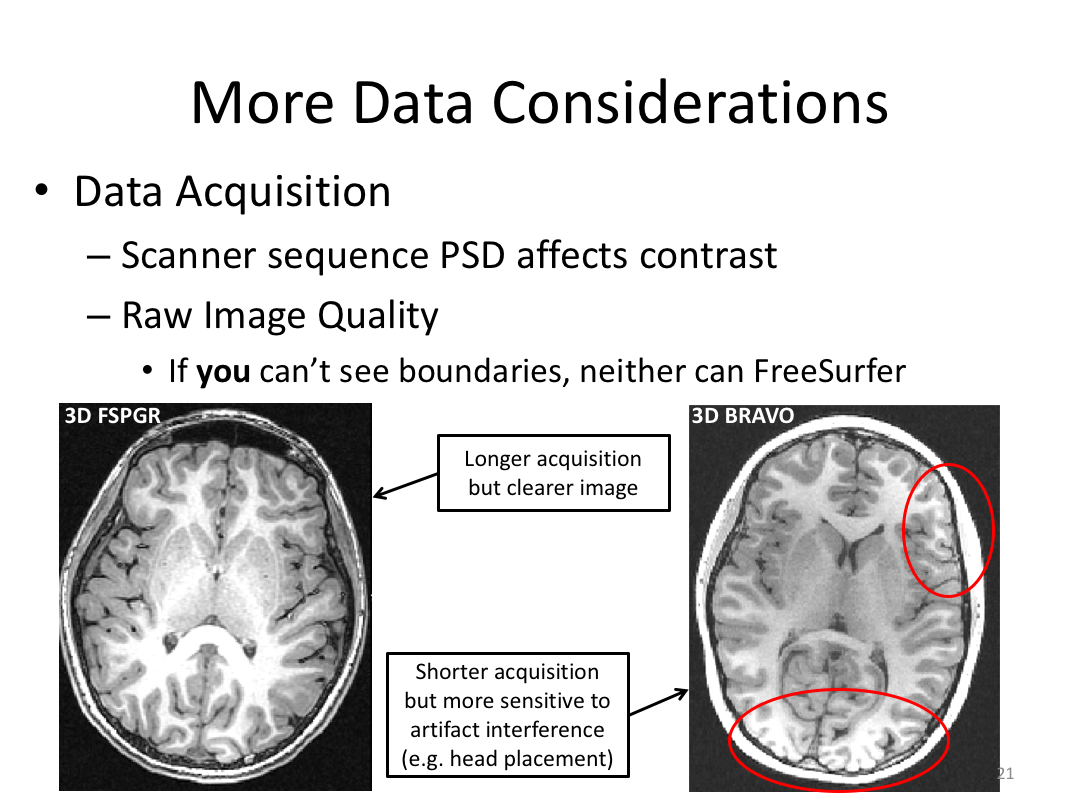


Class 1: MRI Crash Course and Introduction to recon-all
Overview:
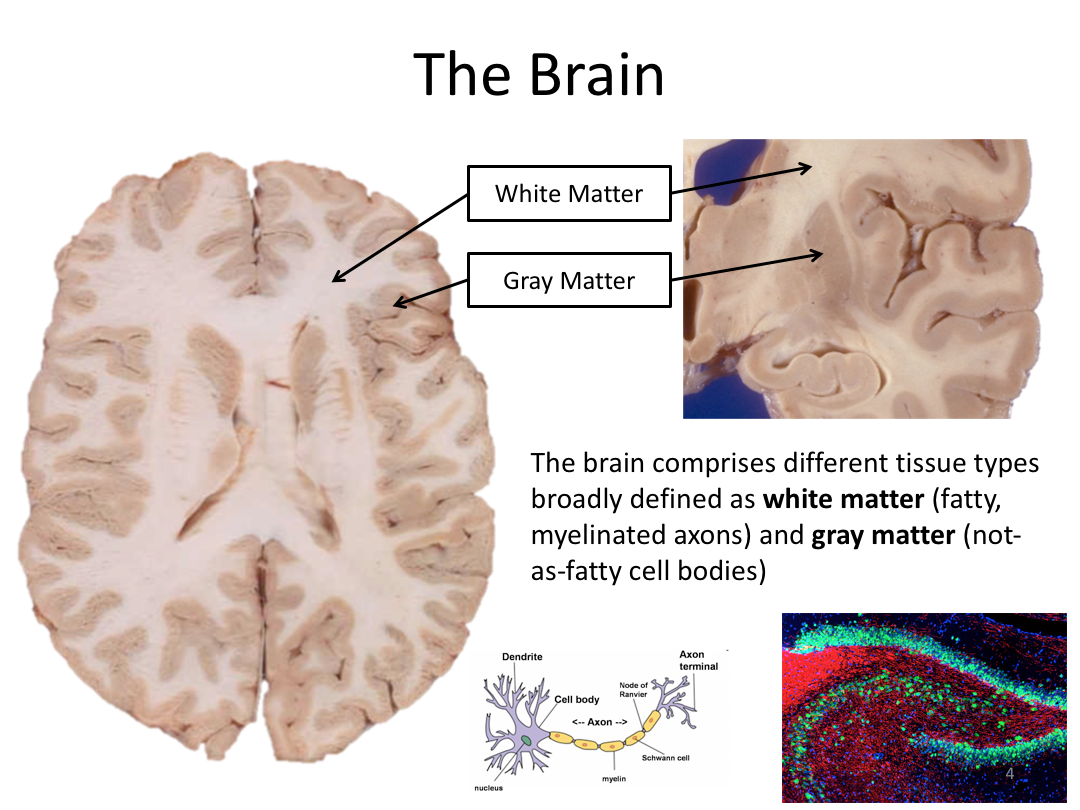
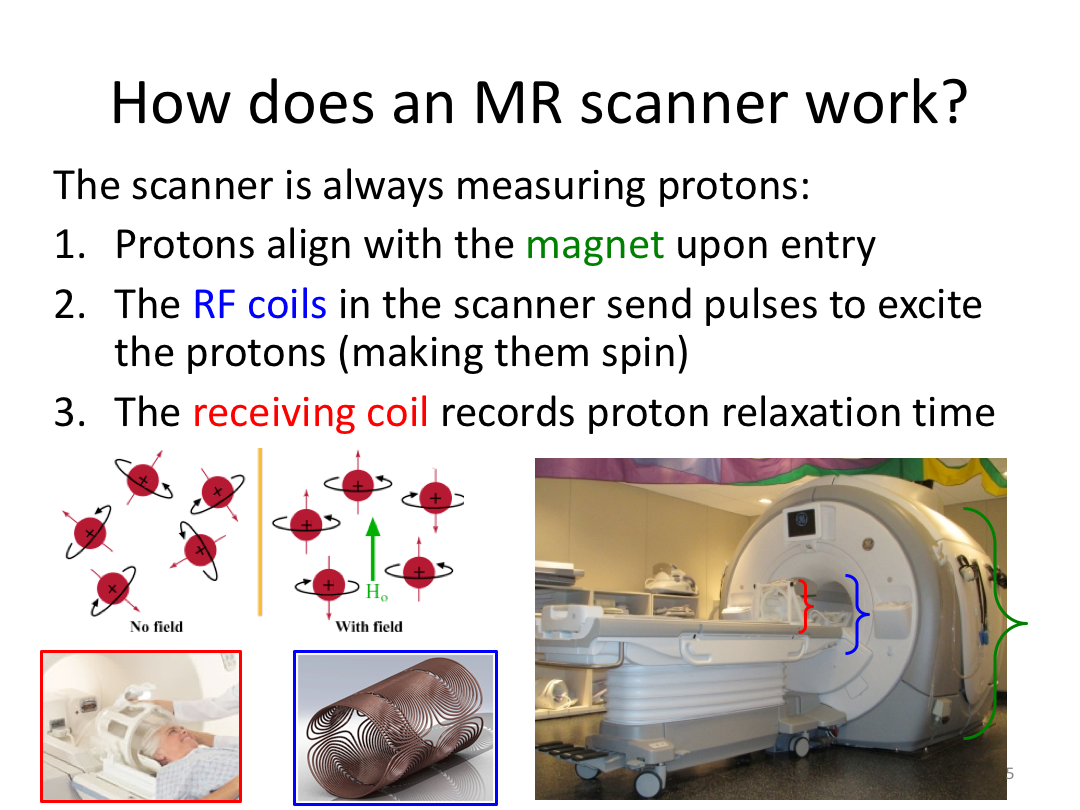
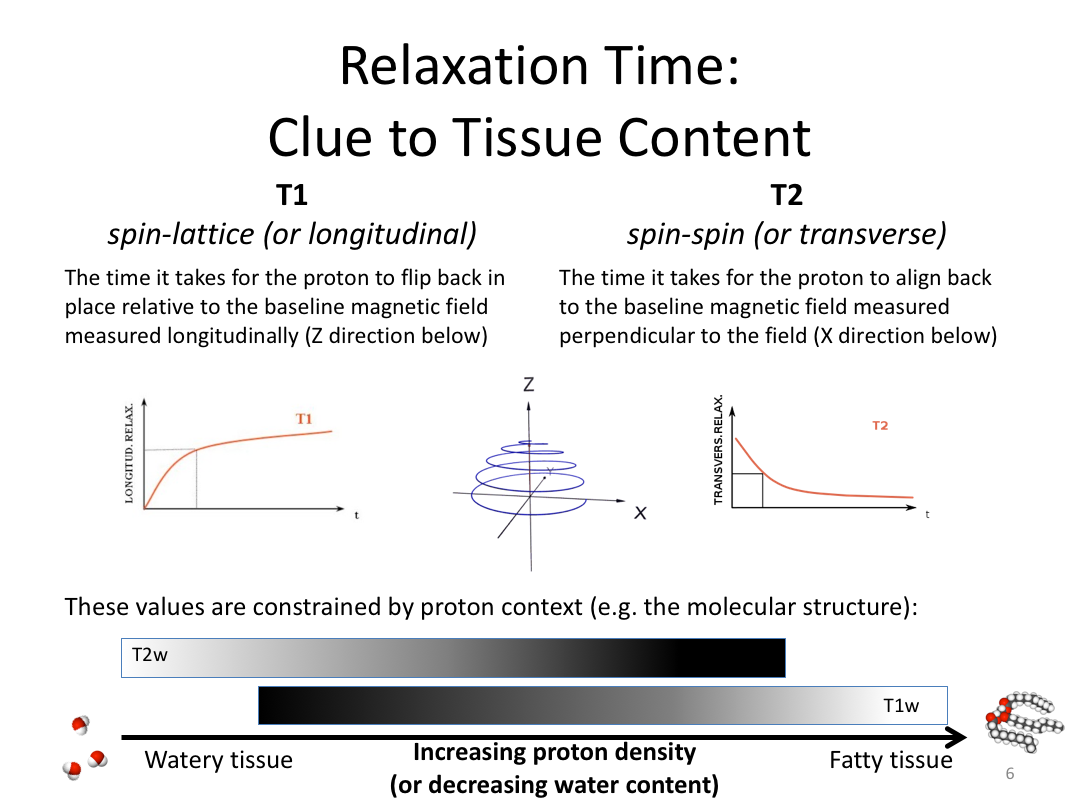
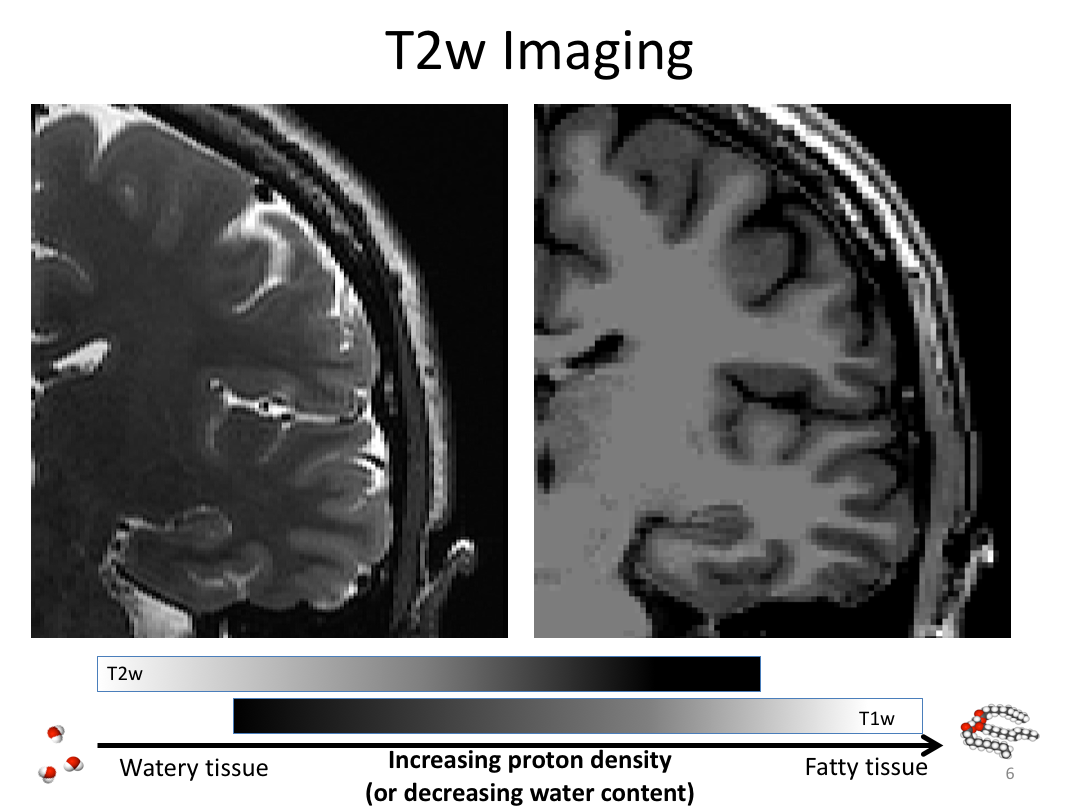
This first class covered the basics of anatomical MR to establish a common understanding of what the scanner is measuring when we collect T1-weighted images. Next, we walked through how to install and navigate FreeSurfer. Finally, we covered how to run recon-all to fully process the practice brains and special considerations when using FreeSurfer for pediatric populations.
Recommended Practice:
1. Install Freesurfer
2. Download the class bin
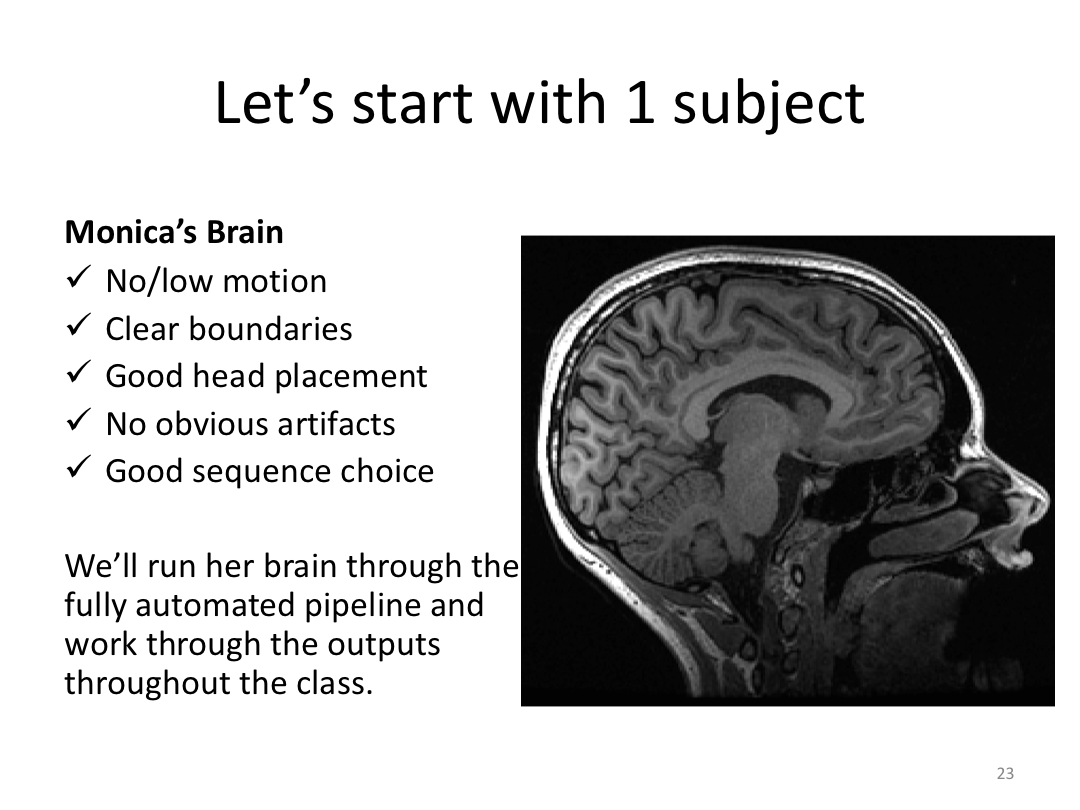
3. Process Monica brain






Class 2: Image Preprocessing (autorecon1)
Overview:
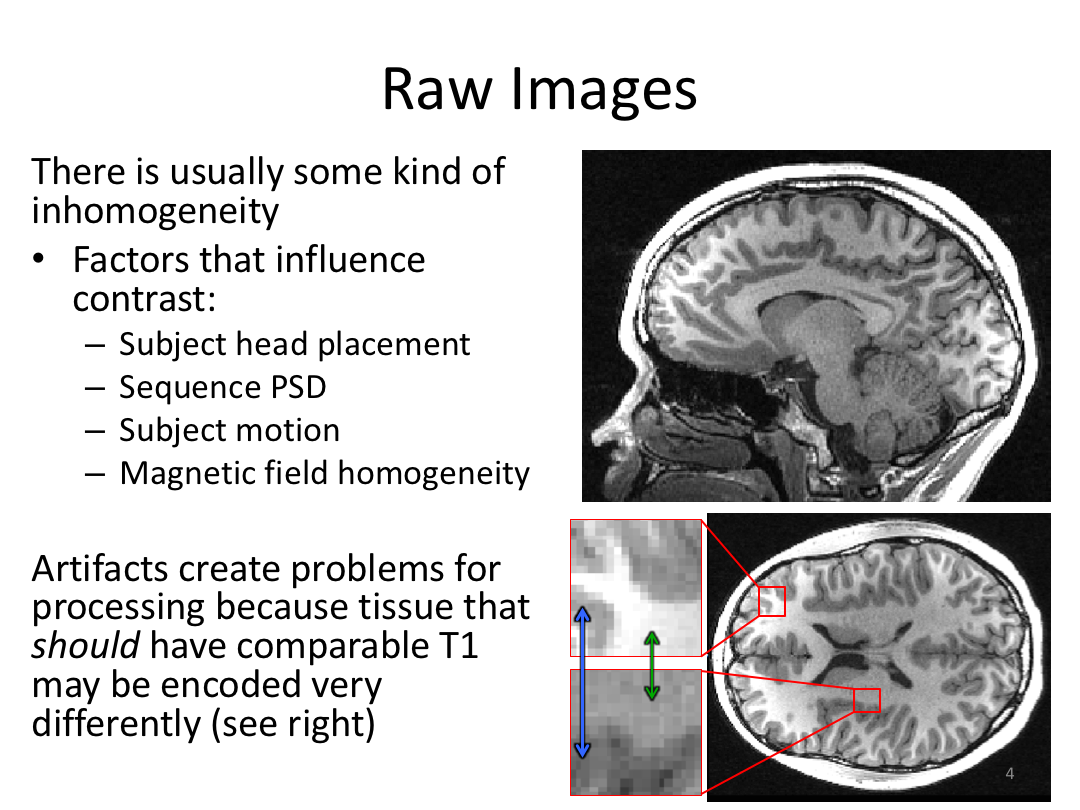
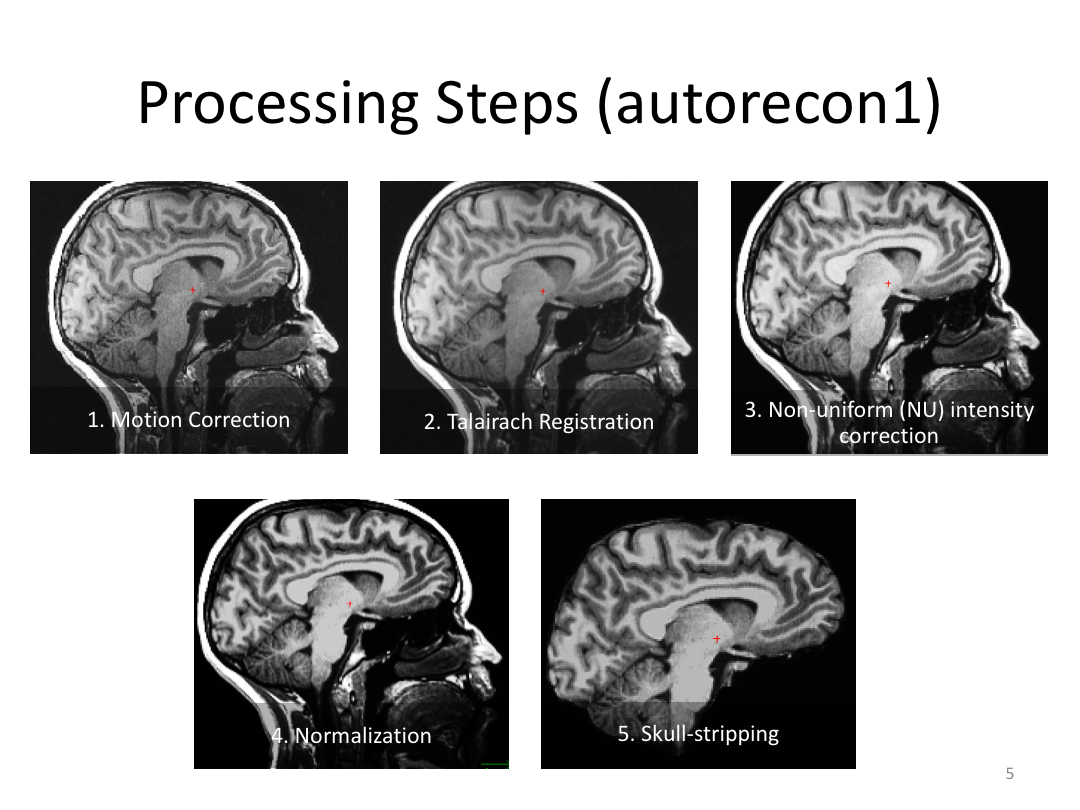
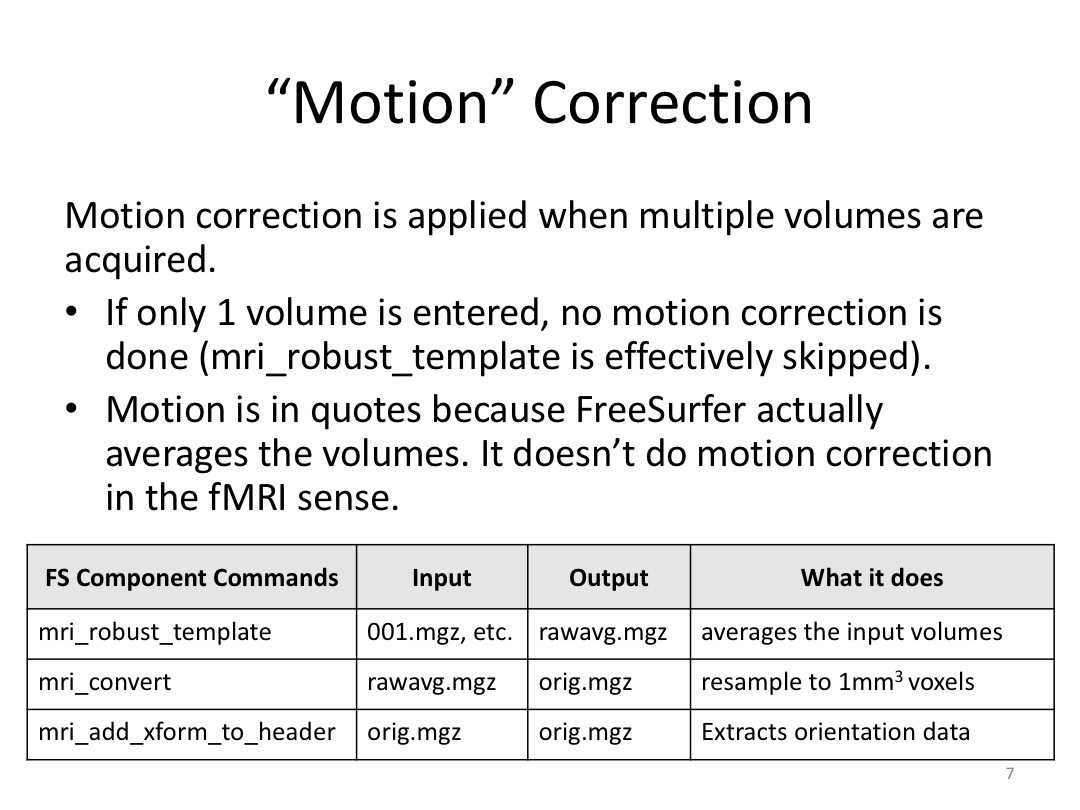
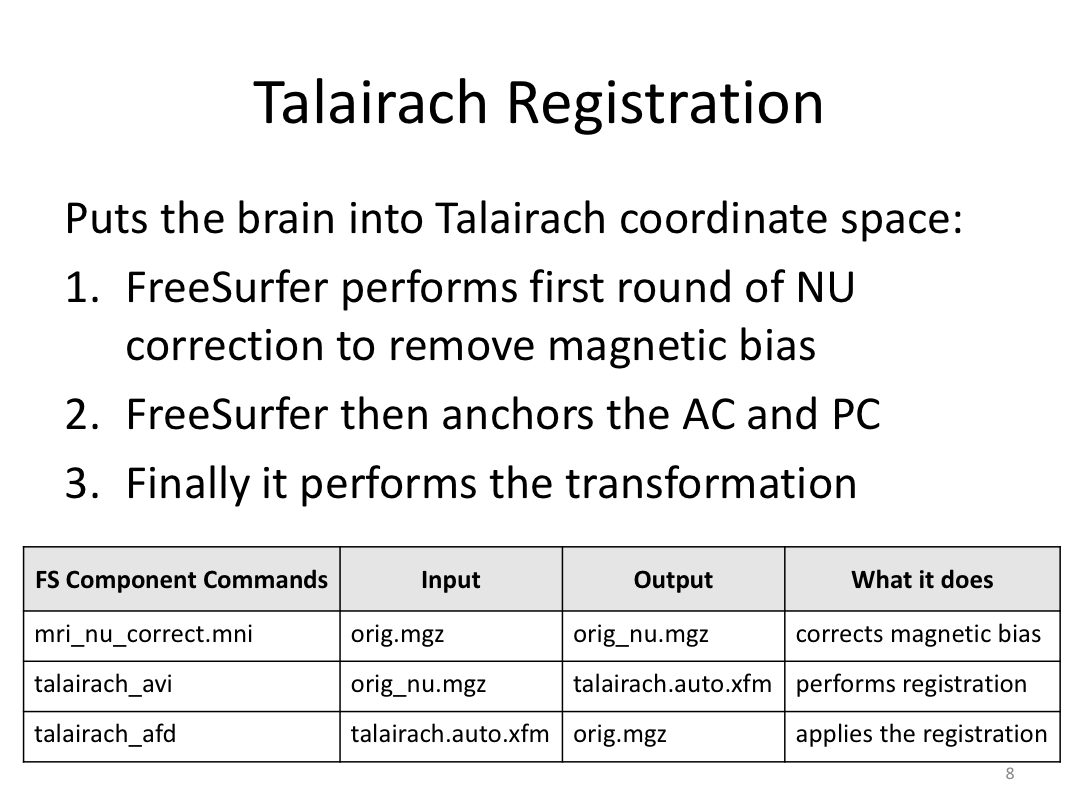
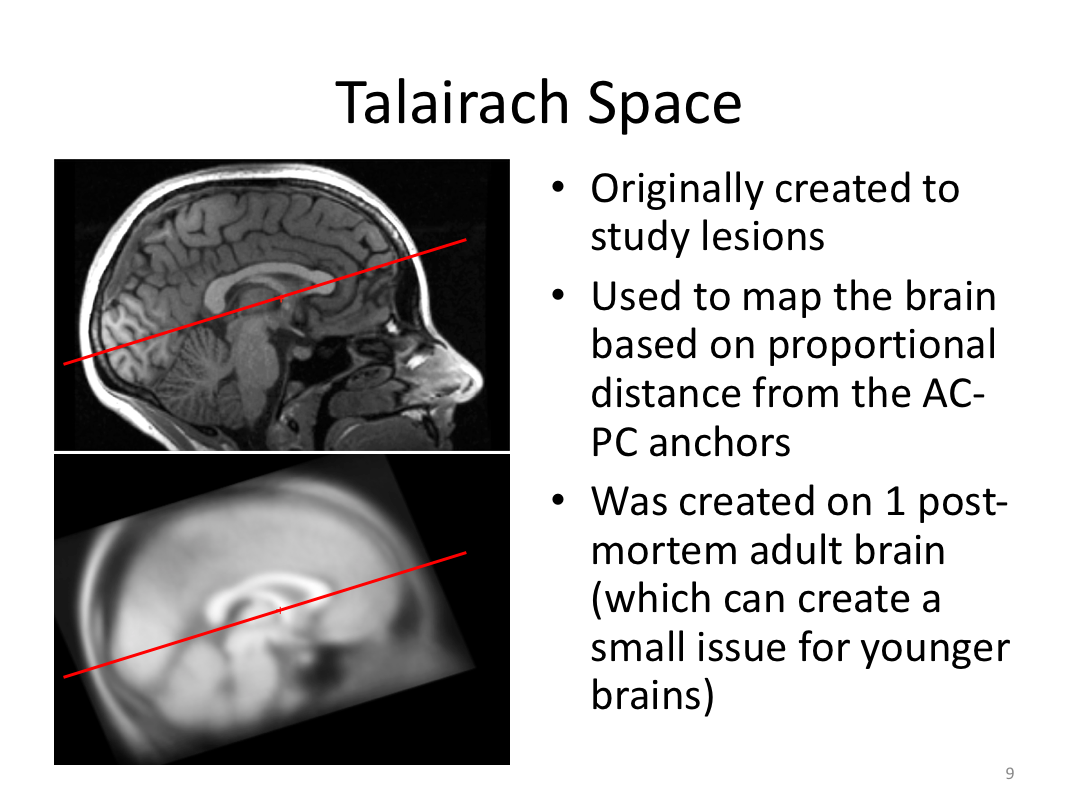
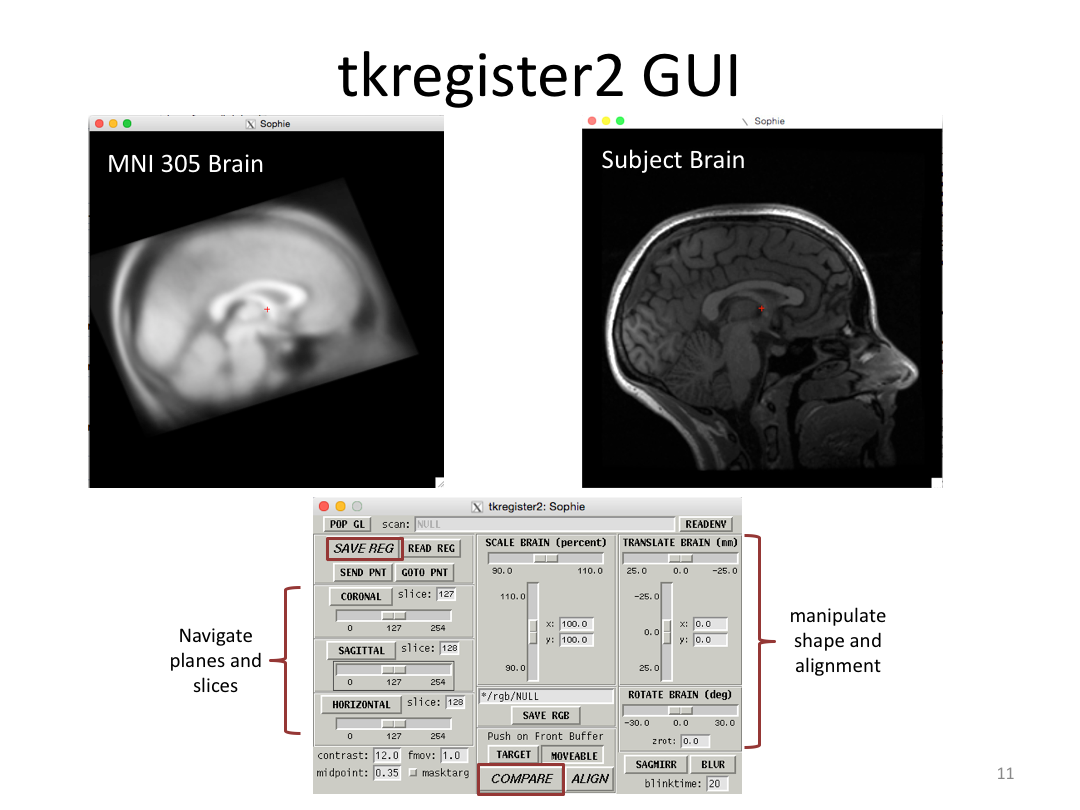
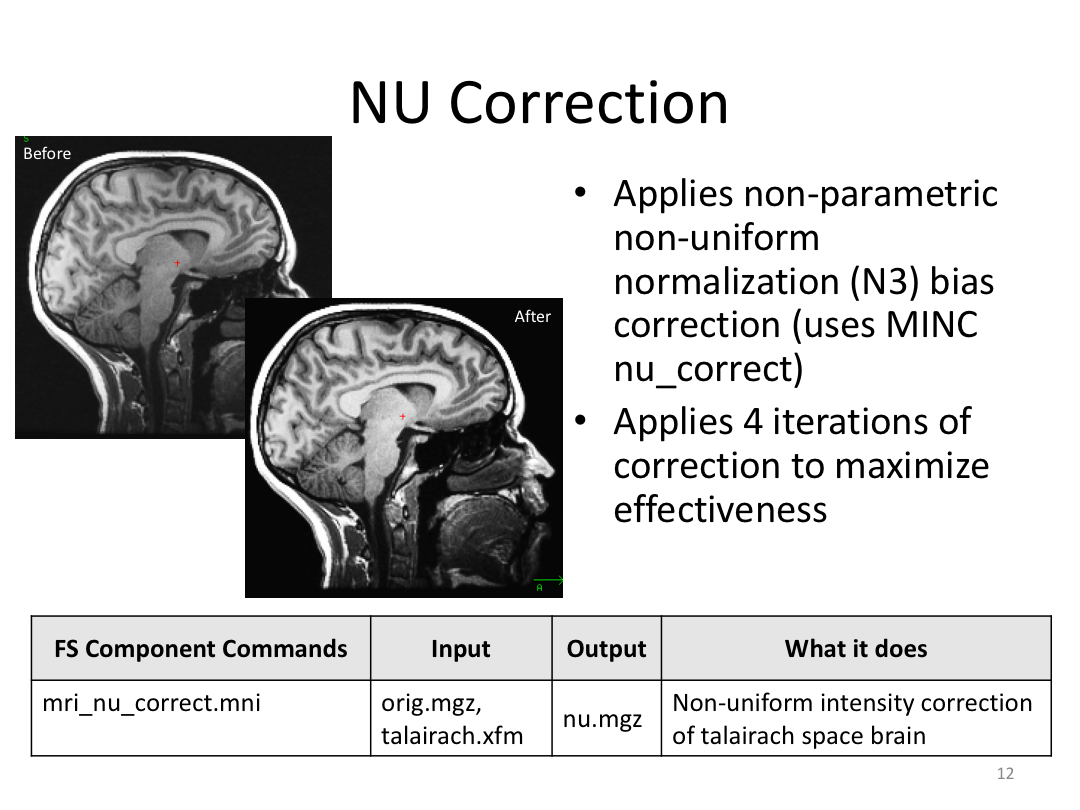
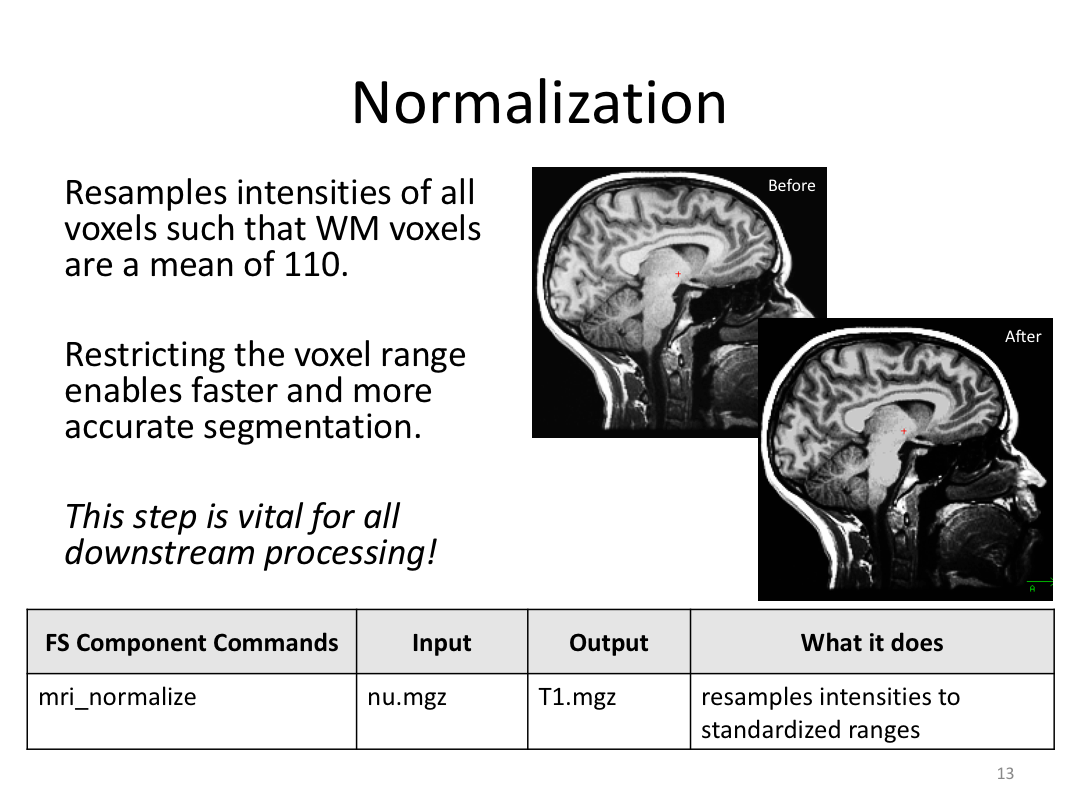
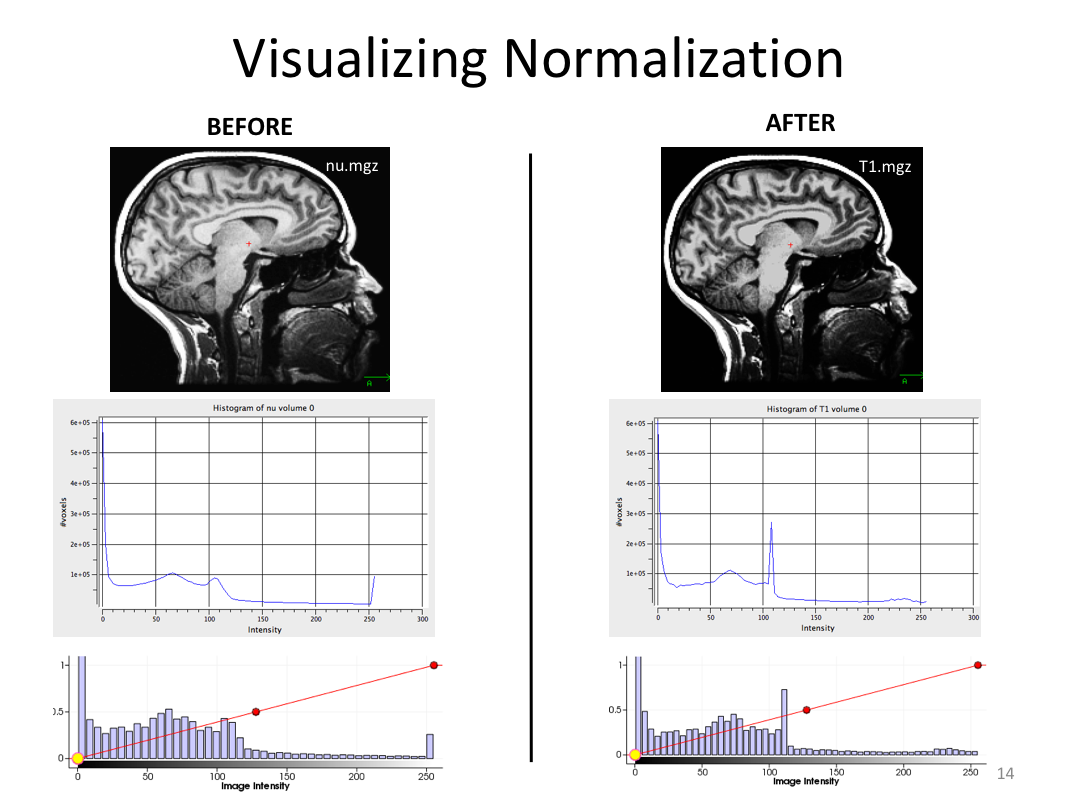
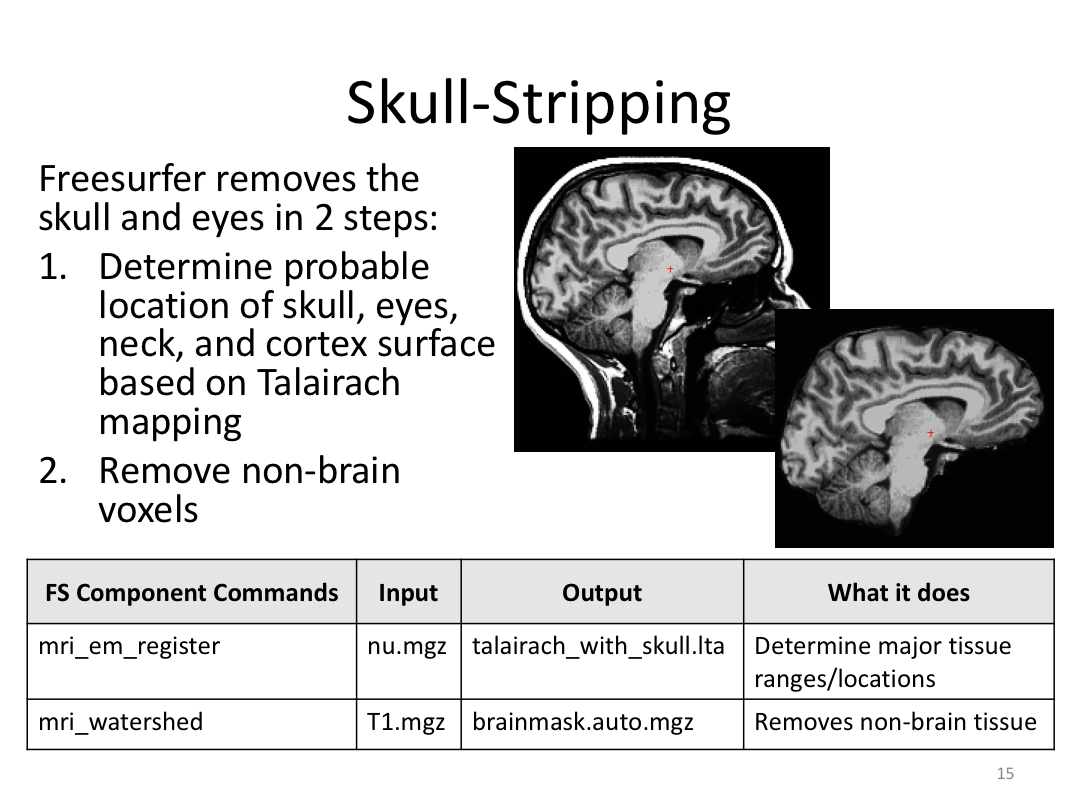
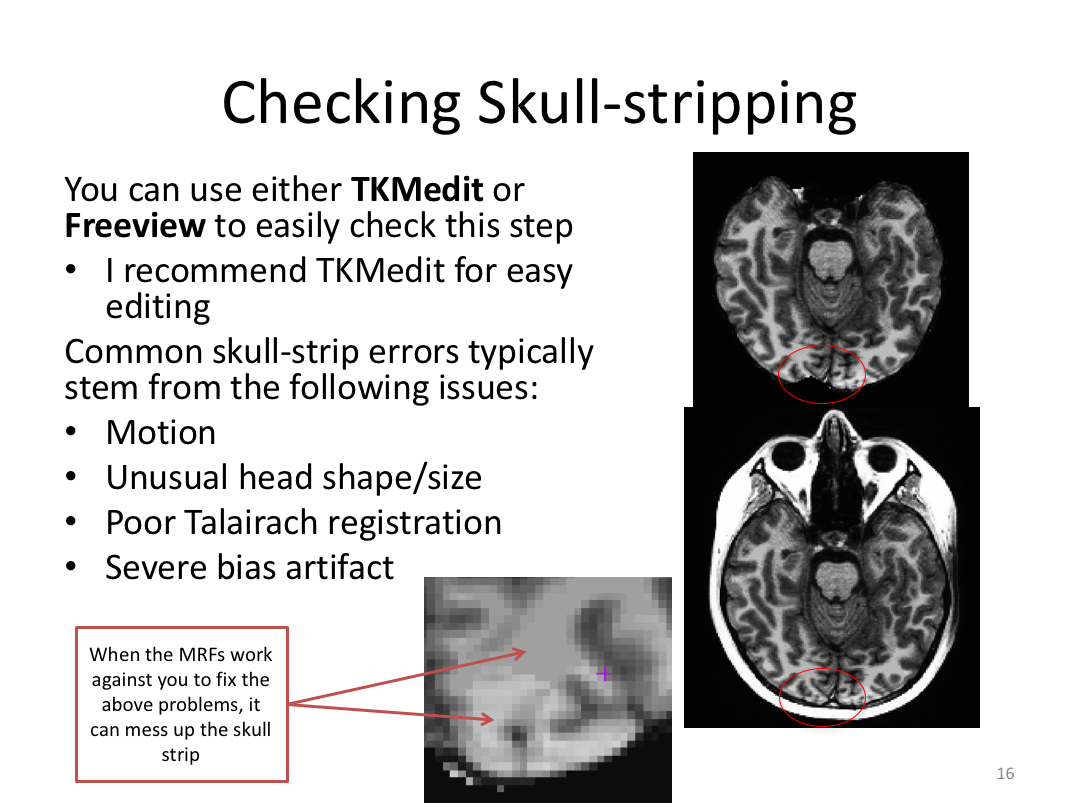
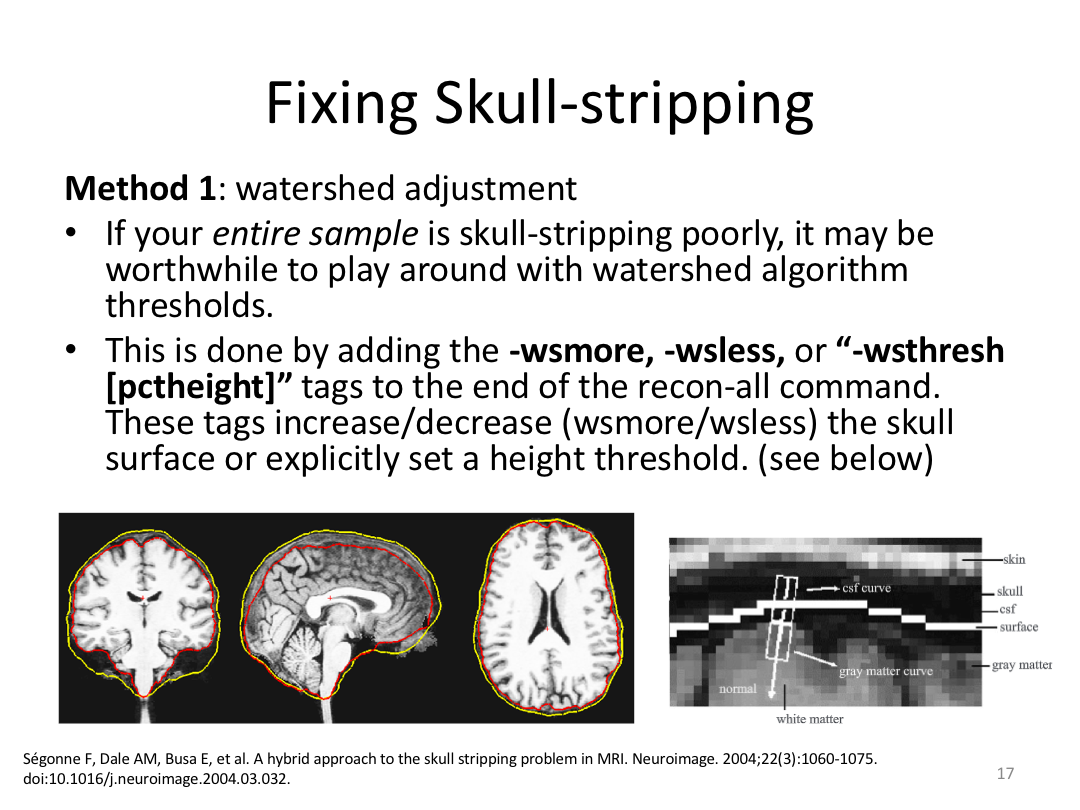
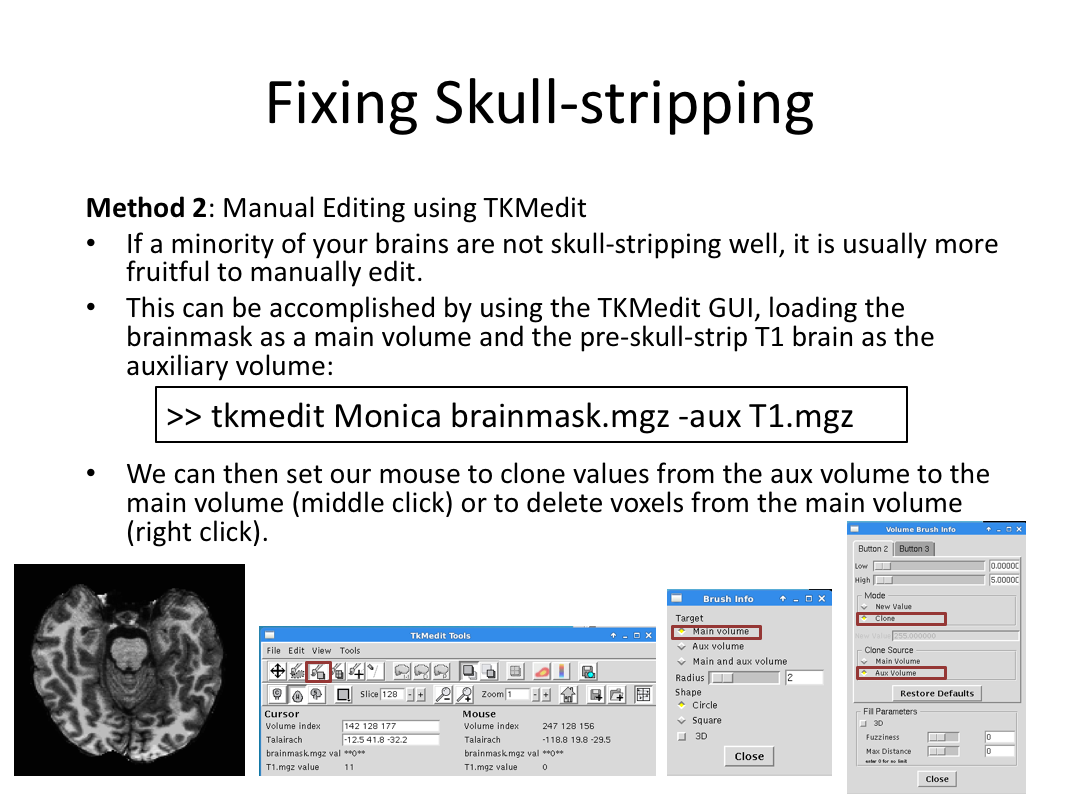
This class covered each step of autorecon1, the initial preprocessing FreeSurfer accomplishes before any segmentation is done. This includes 1) Talairach registration, 2) intensity normalization, and 3) skull stripping. This class also introduces the TKregister2 tool to fix registration issues and the TKMedit interface to fix issues with skull stripping.
Recommended Practice:
1. Process Meghan brain
2. Manually adjust Talairach registration
3. Edit Monica or Meghan's brainmask

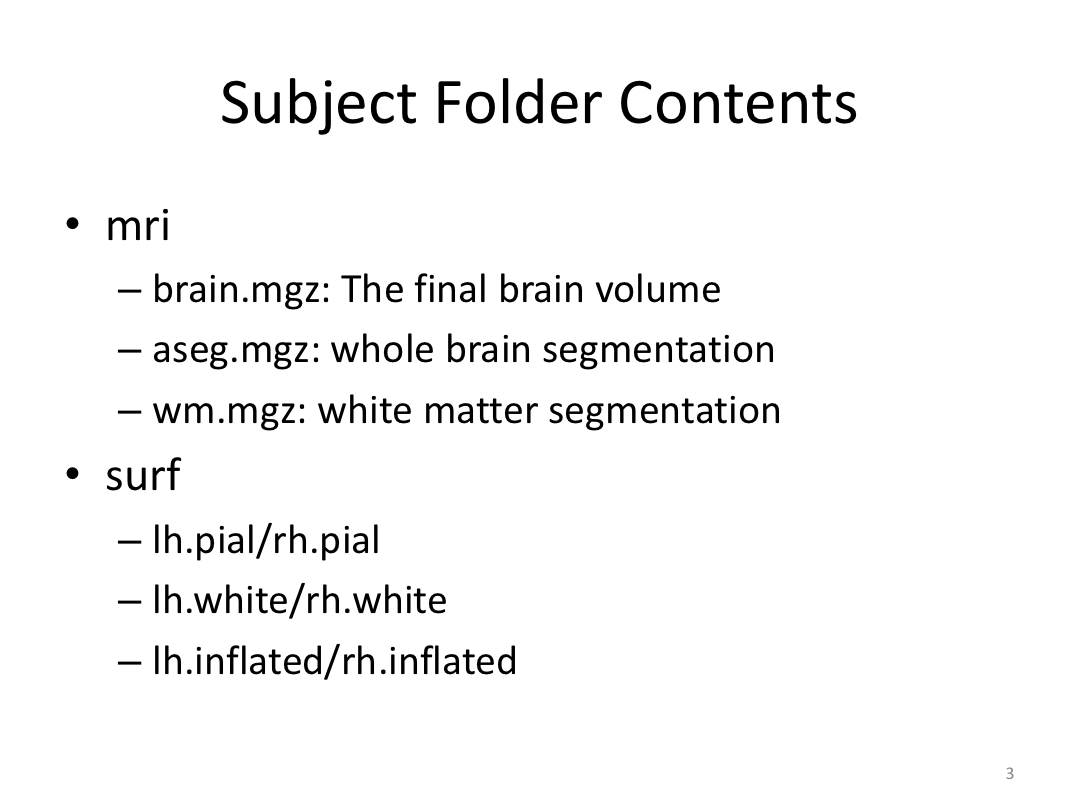
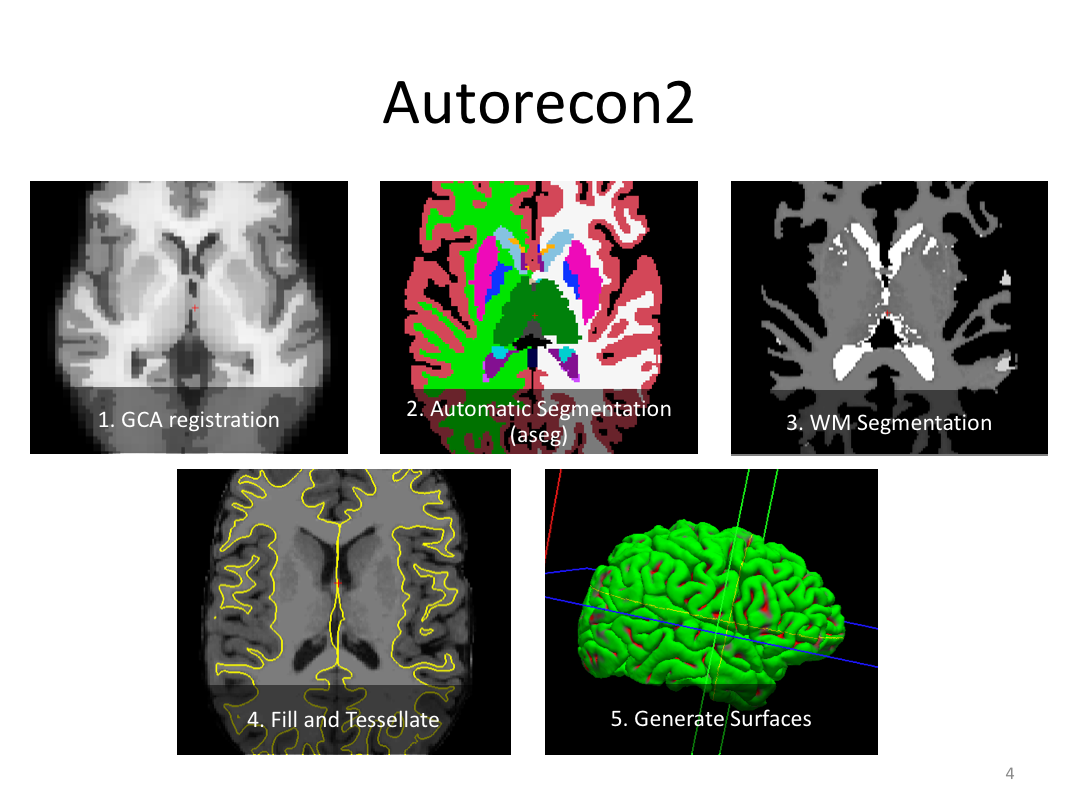

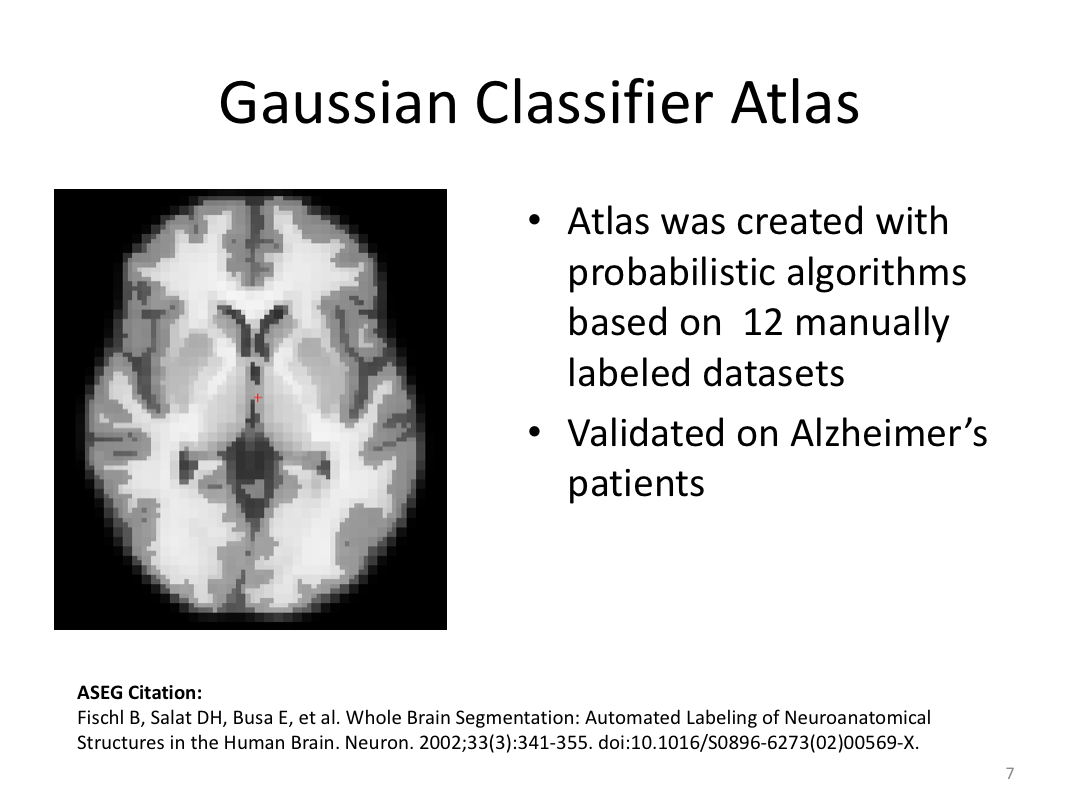
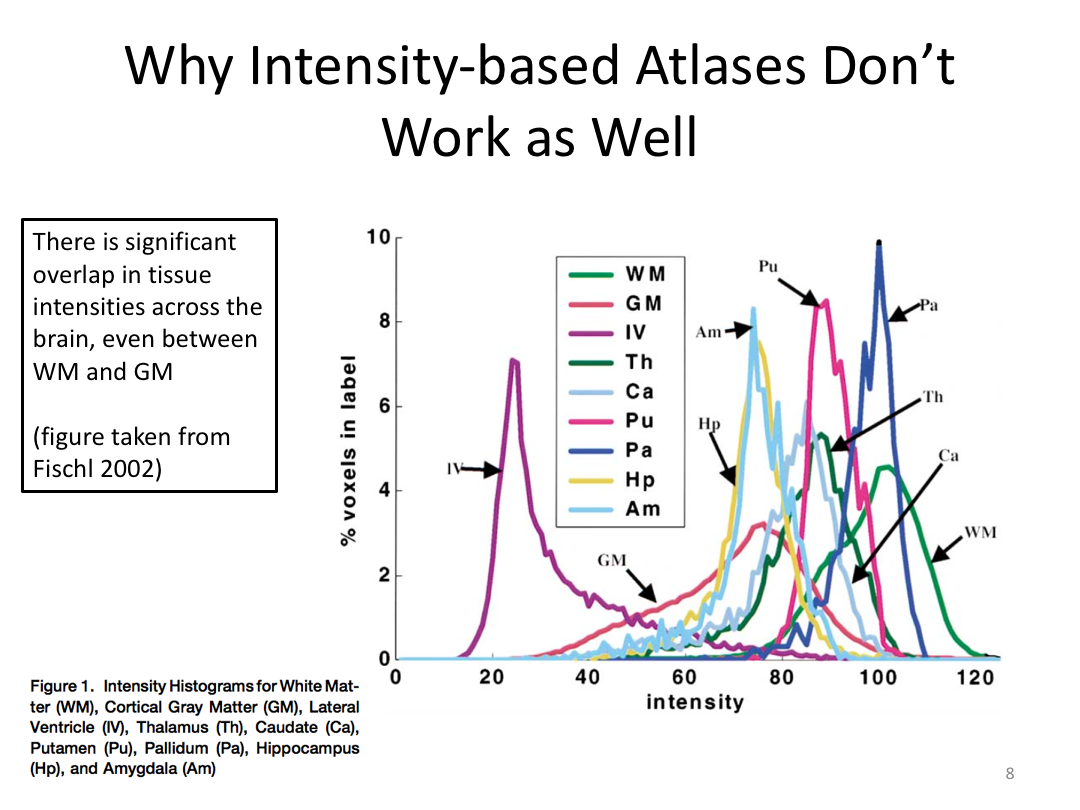
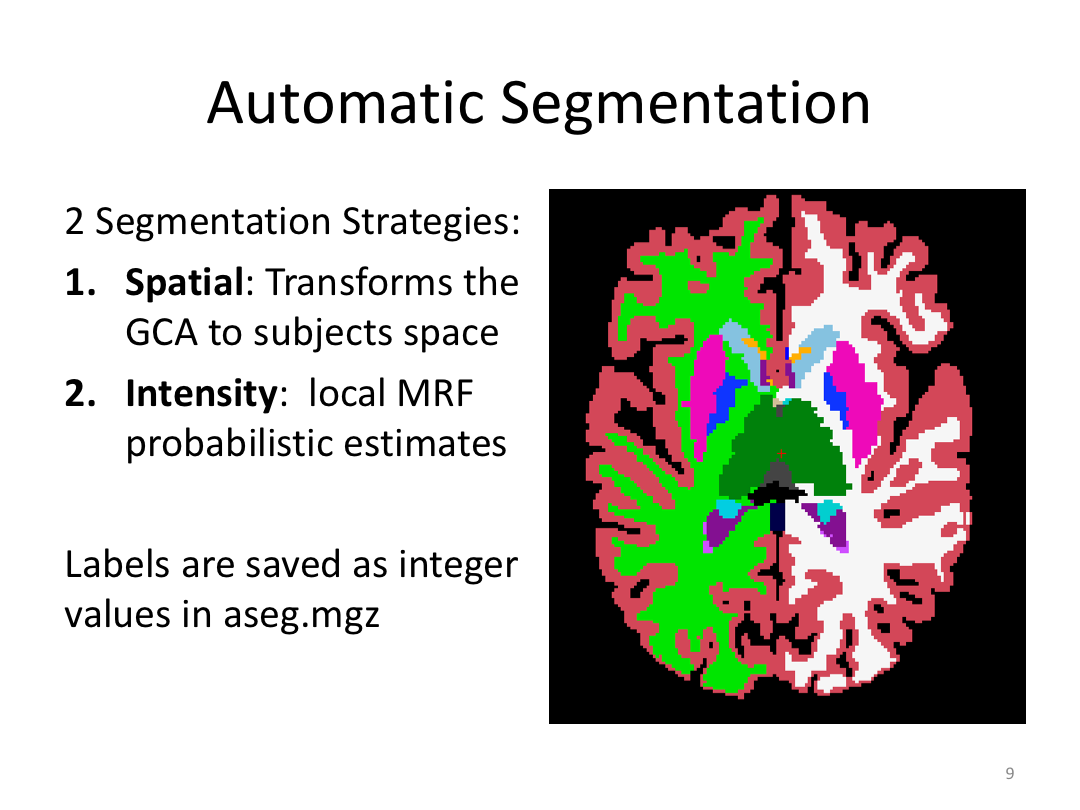
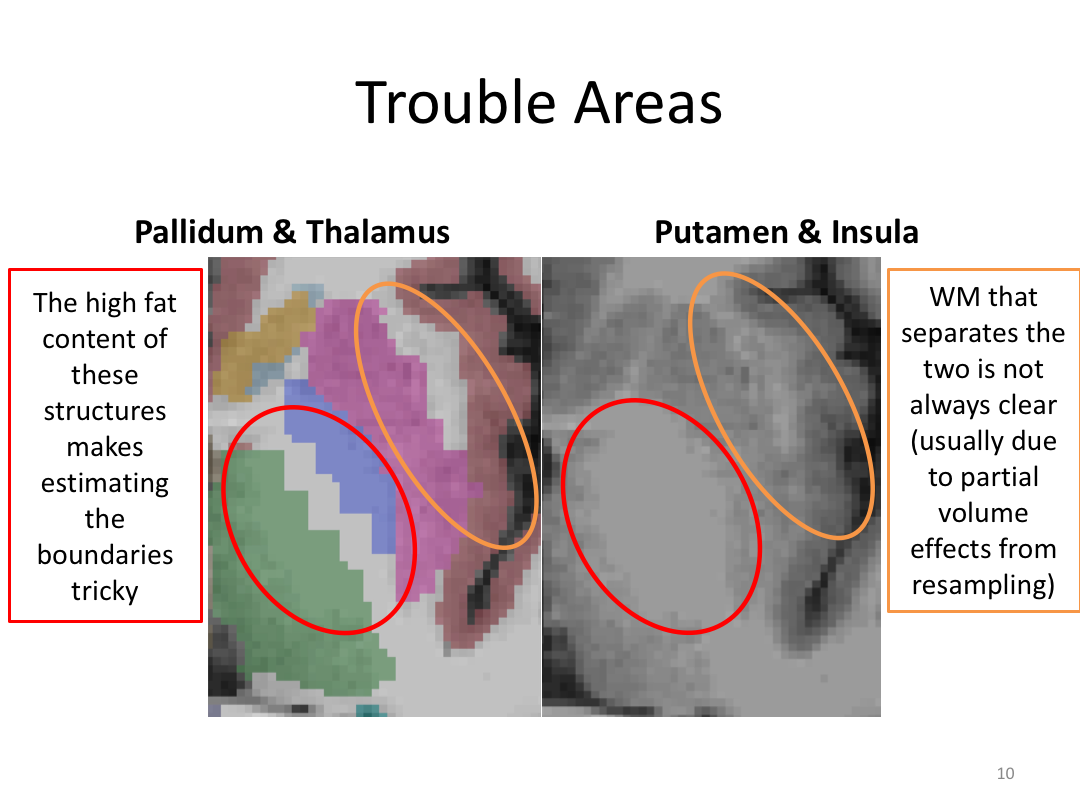
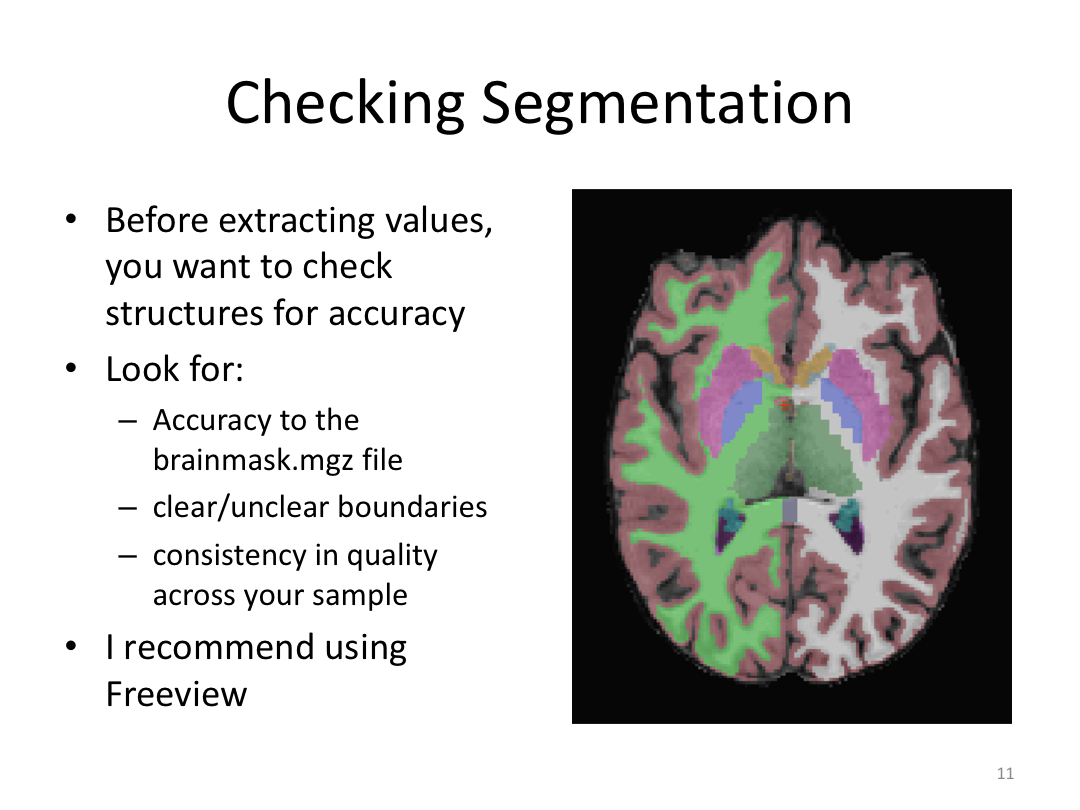
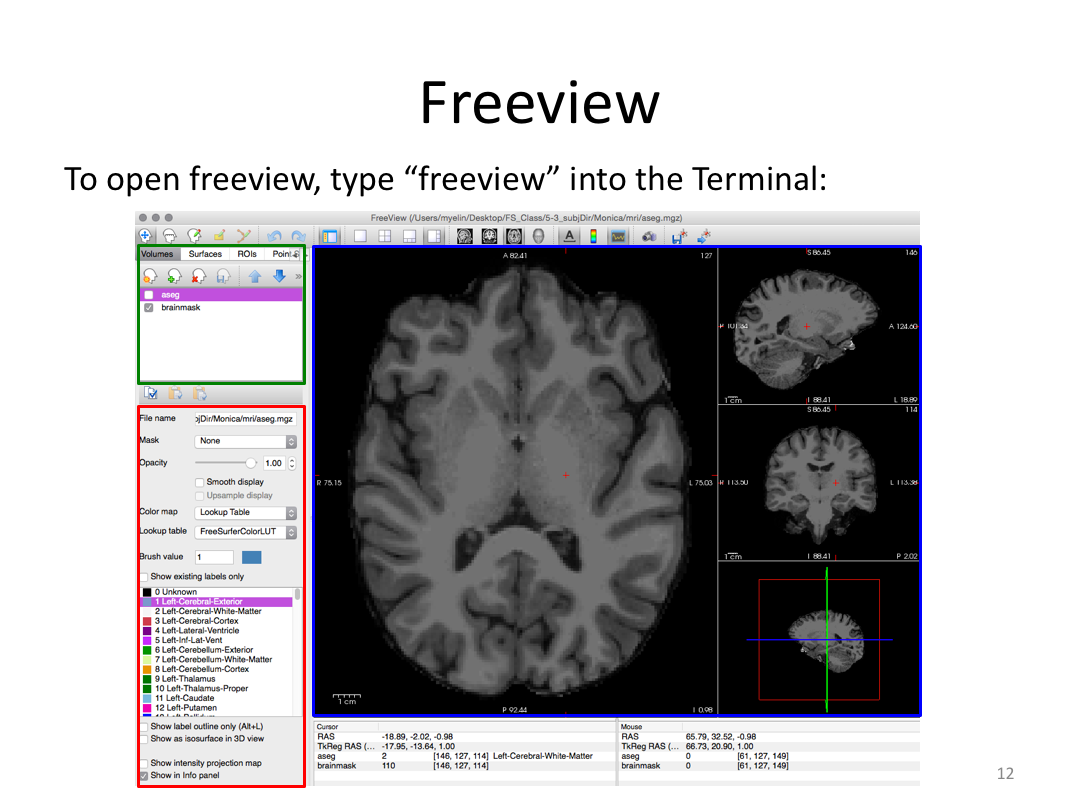
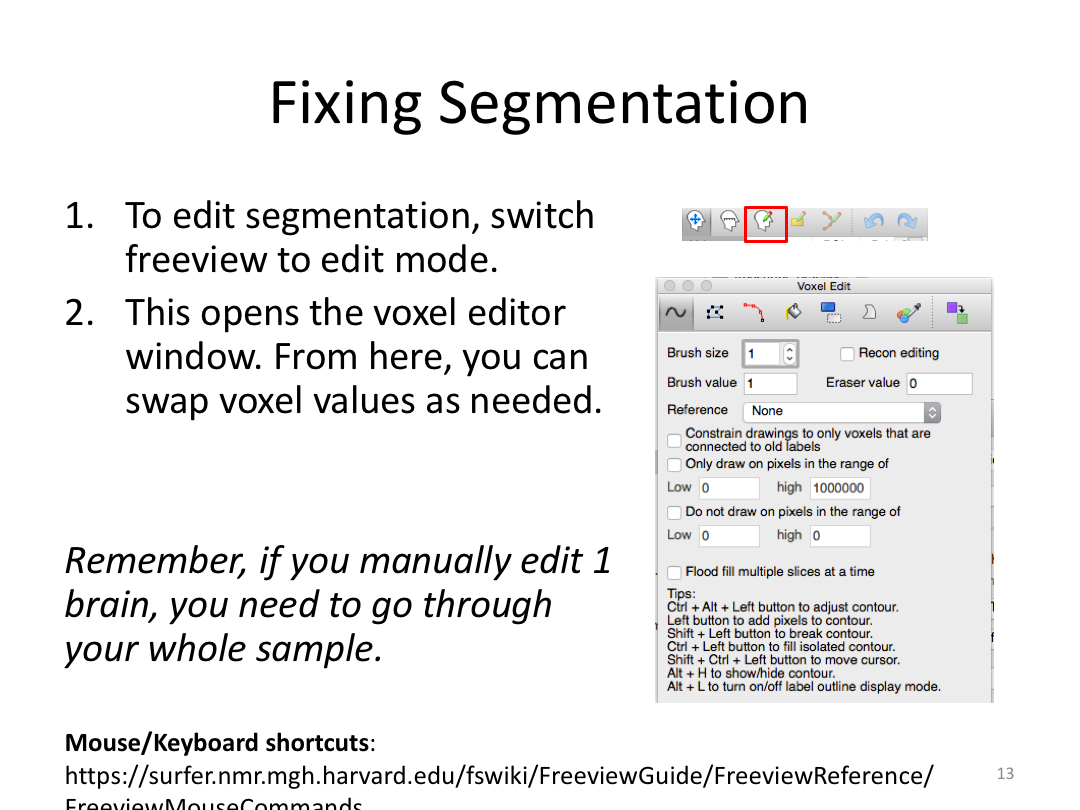
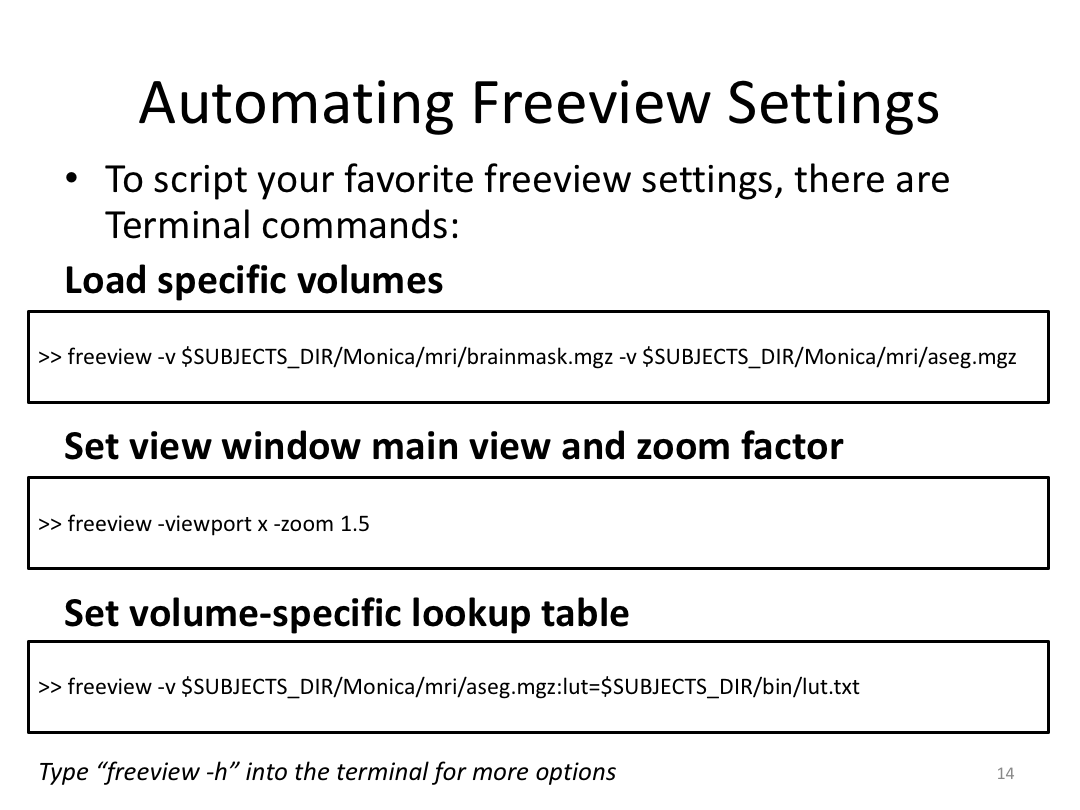
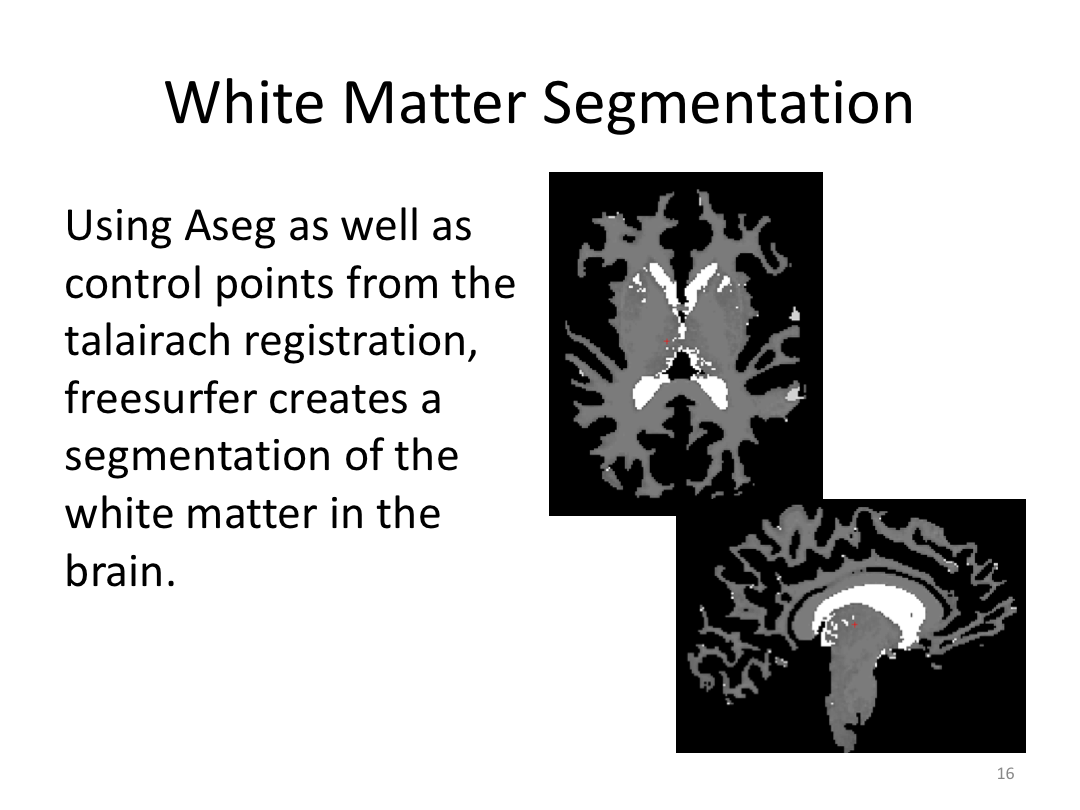
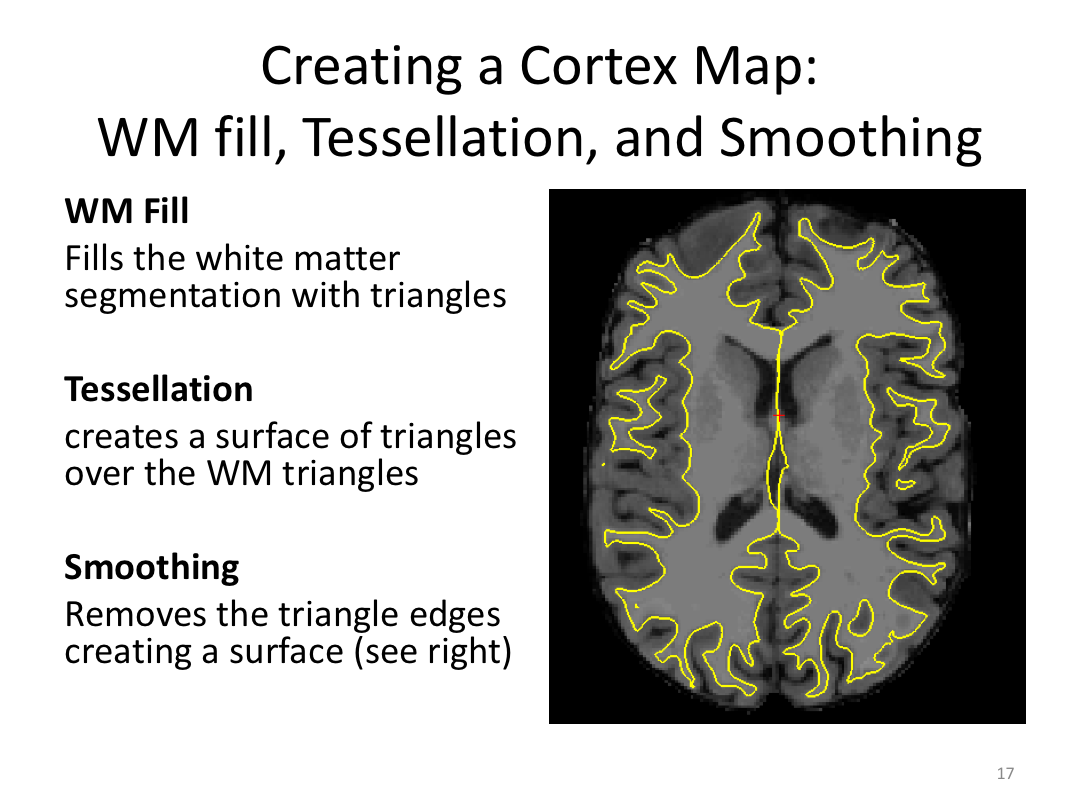
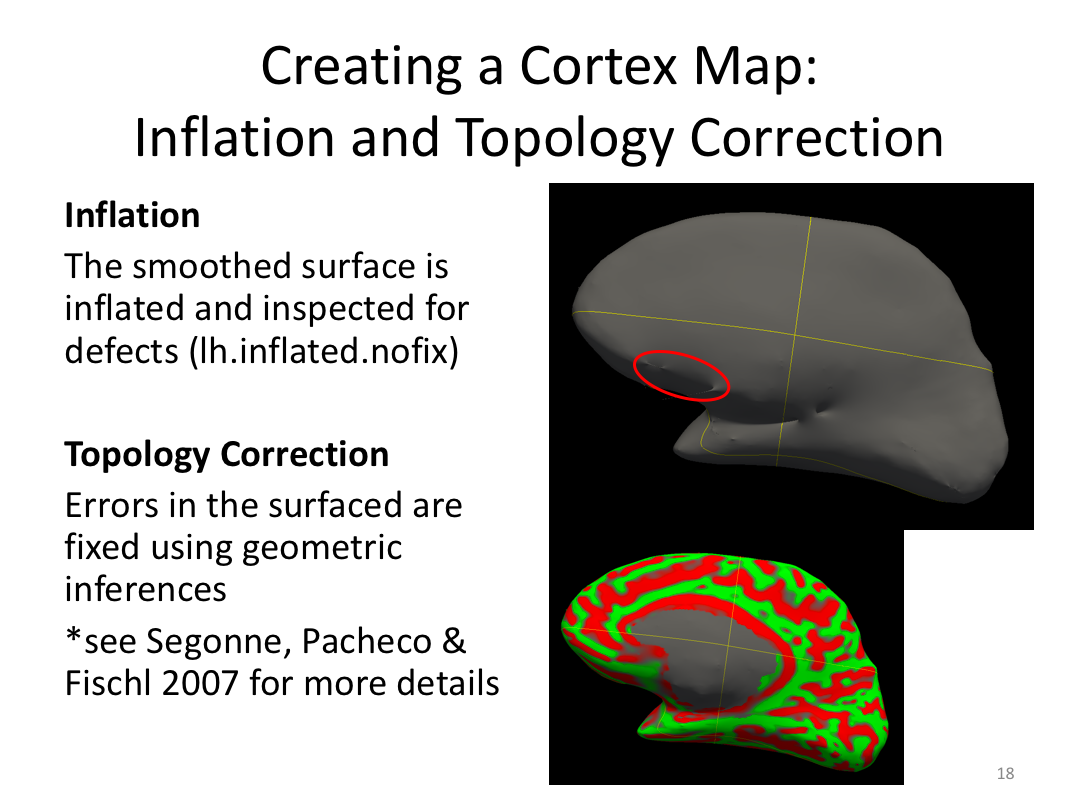
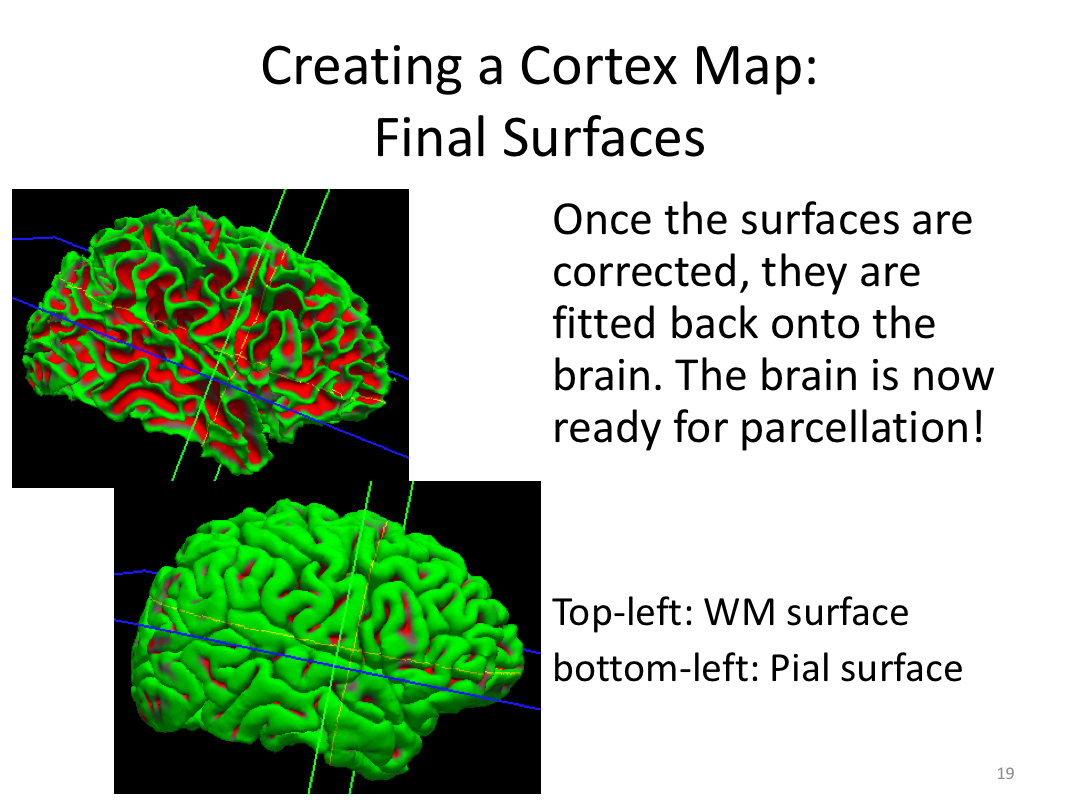
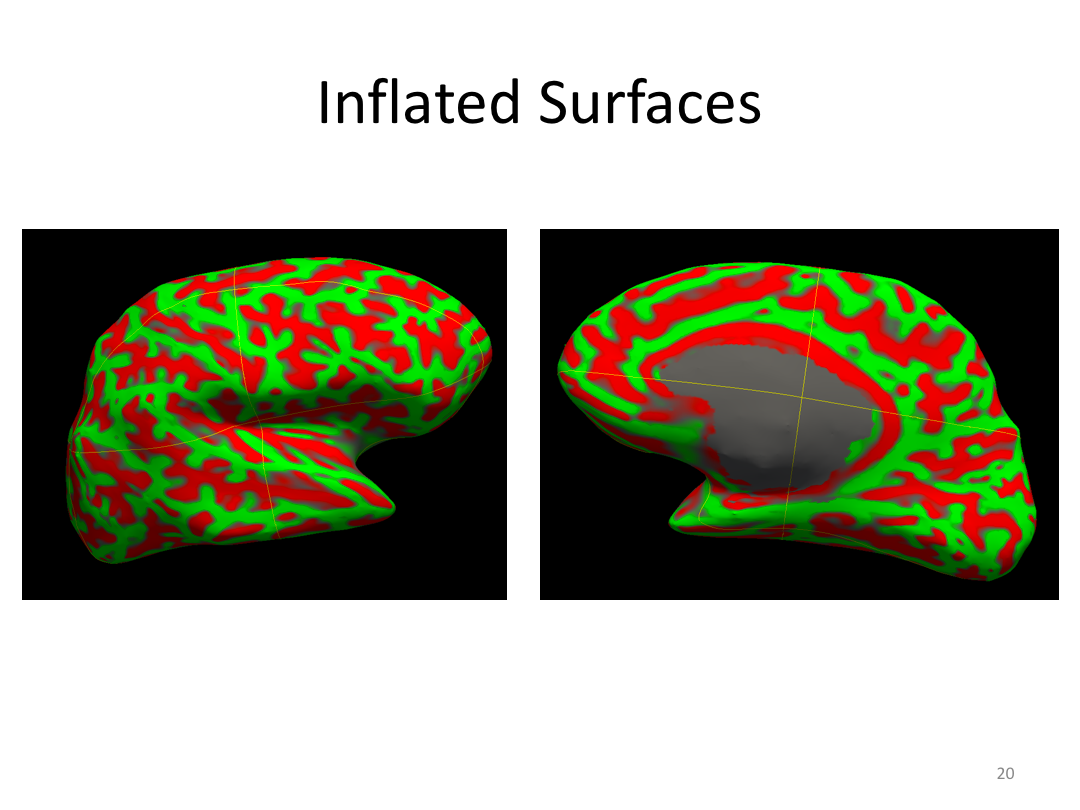

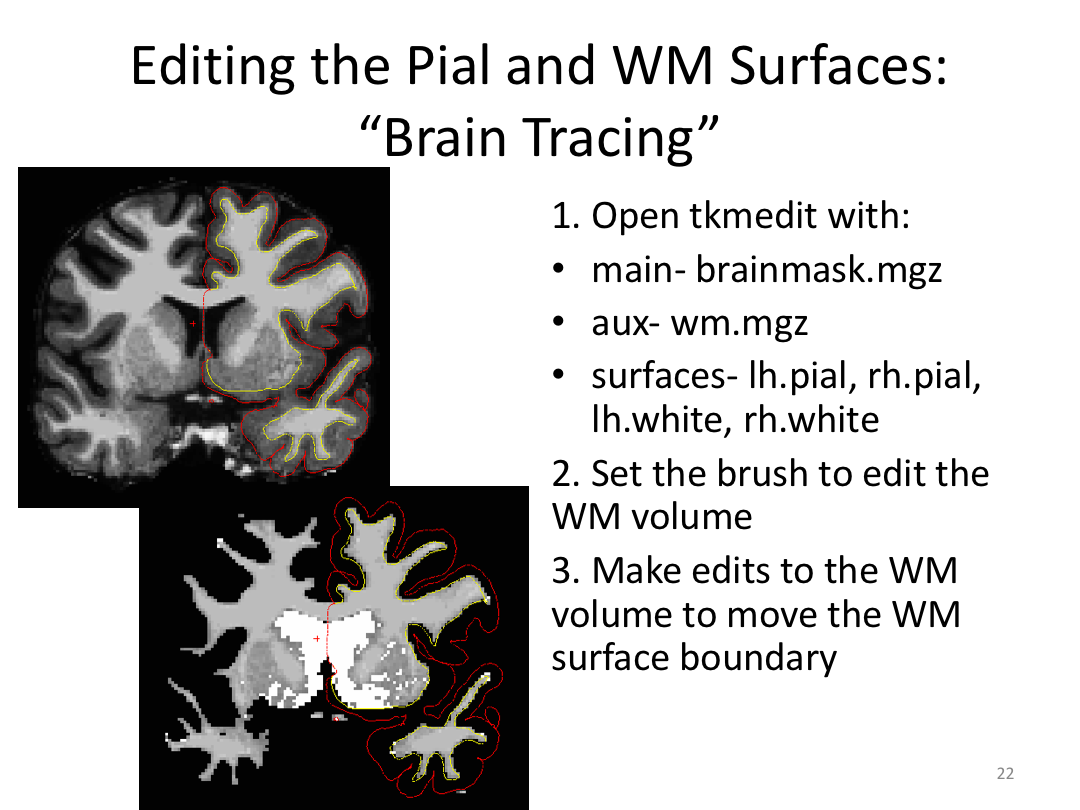
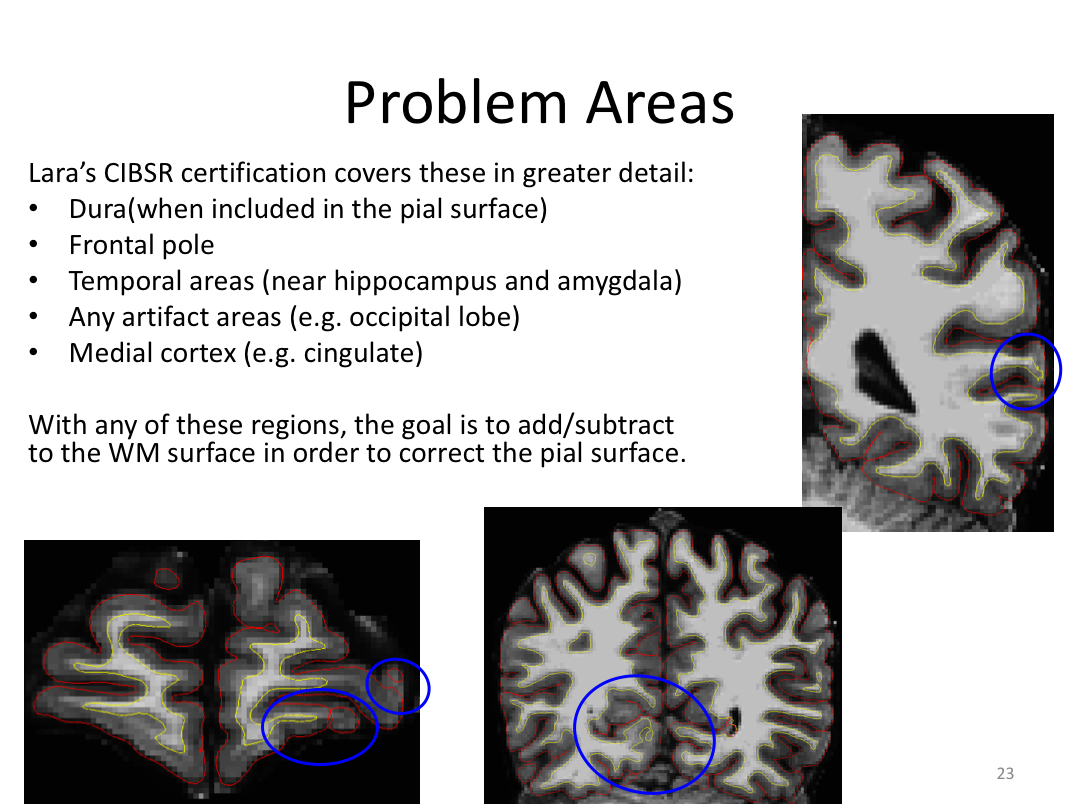
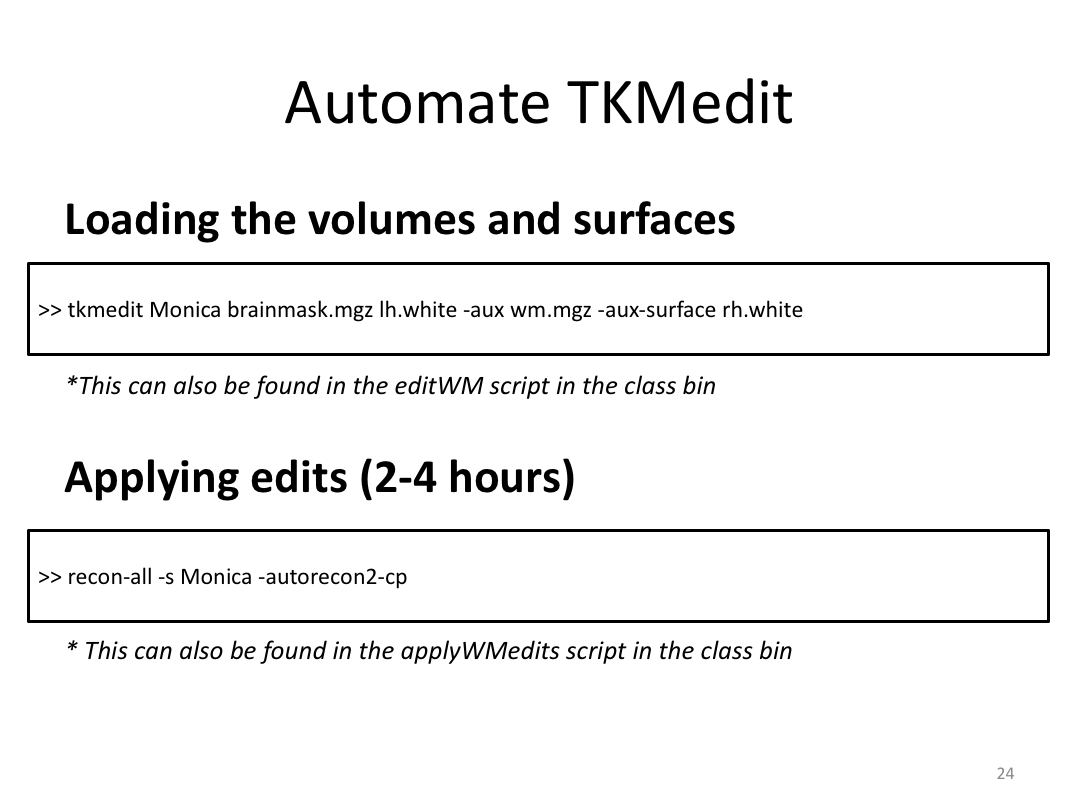

Class 3: Automatically Segmenting the Brain (autorecon2)
Overview:
This class covered the two major components of autorecon2: automatic segmentation and surface generation. We covered the steps taken to accomplish each and introduced quality control steps that can be implemented for both subcortical volumes and for cortical surfaces (before parcellation; covered in class 4). Surface editing introduced another use of the TKMedit GUI, and we introduced the FreeView GUI for segmentation quality checking and for manually editing segmentation.
Recommended Practice:
1. Run a brain through autorecon2
2. Pull up the wm.mgz and brainmask.mgz files in TKMedit
3. Edit the pial/wm surface boundaries
4. Open the aseg.mgz and brainmask.mgz files and scroll through the brain


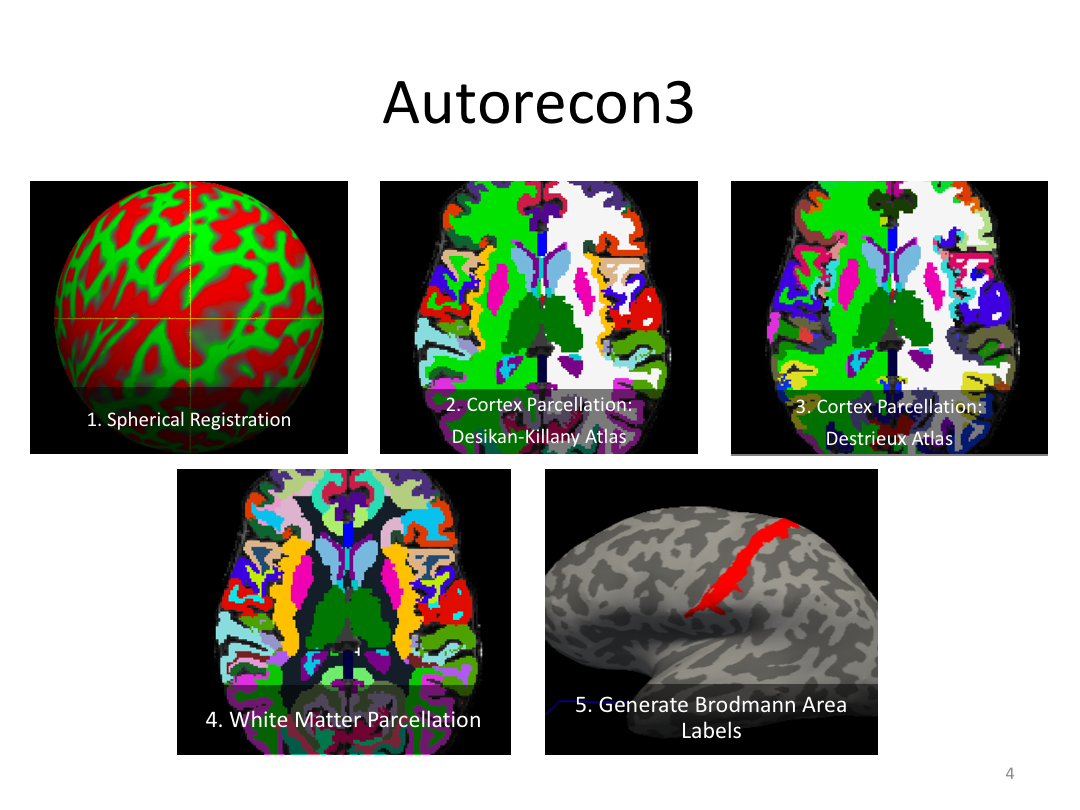

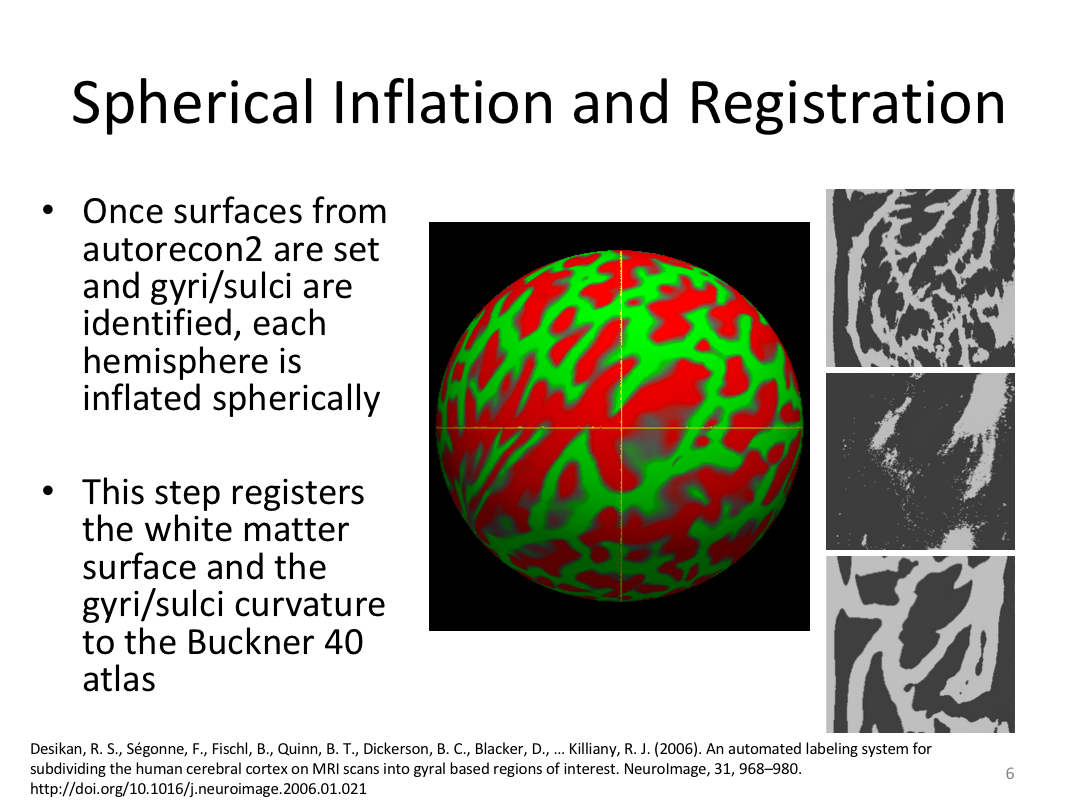
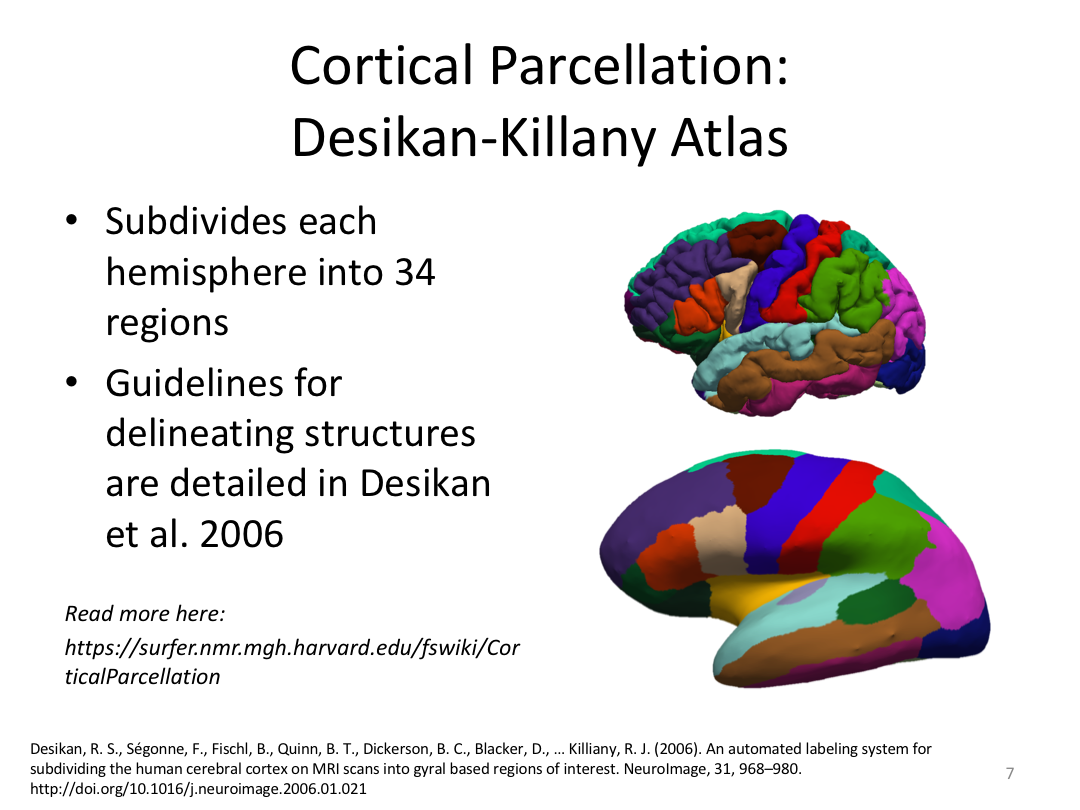
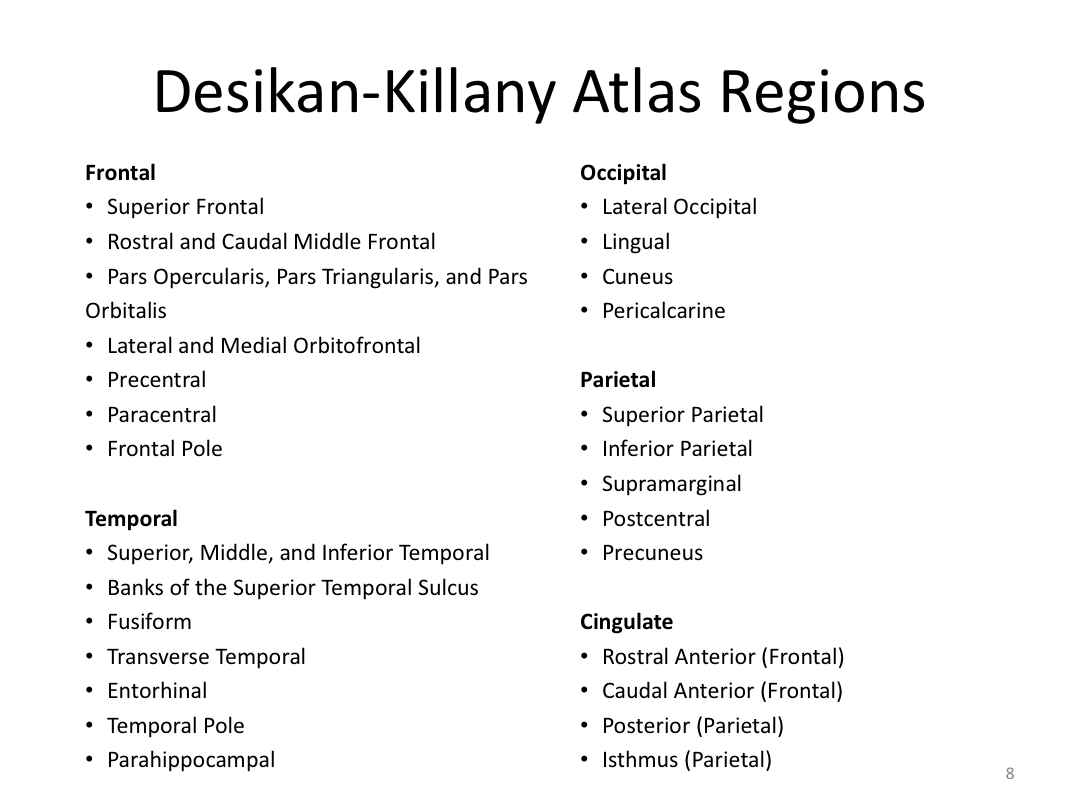
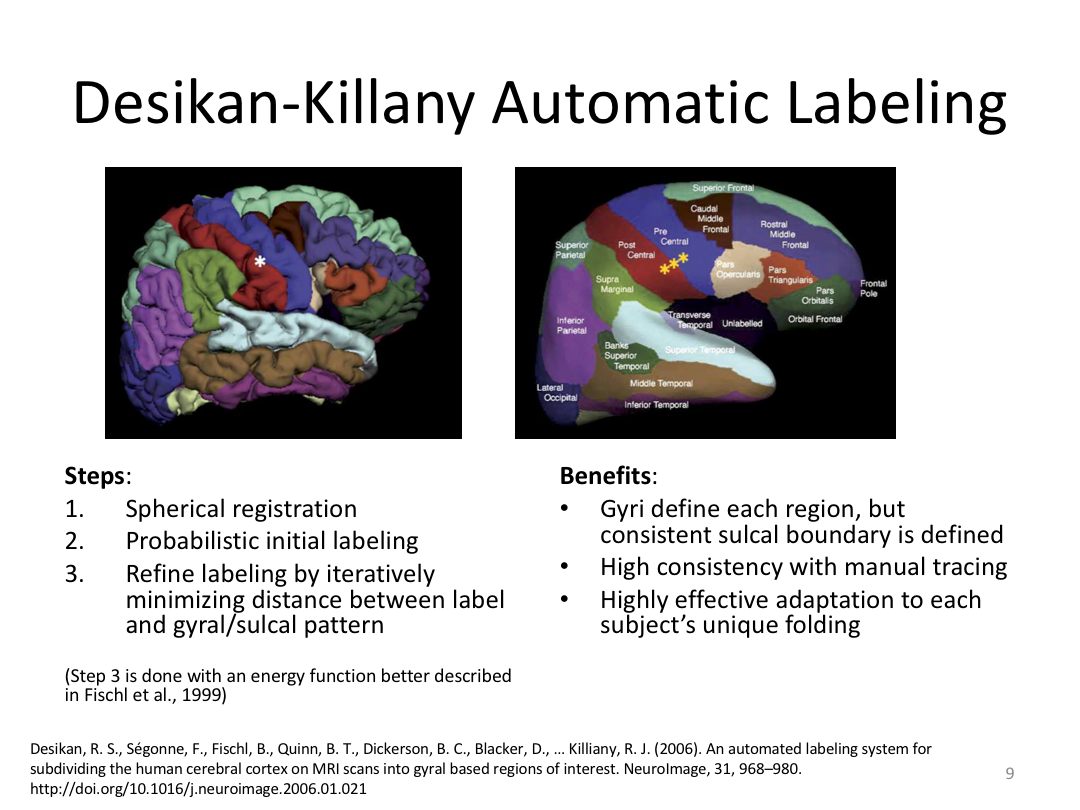
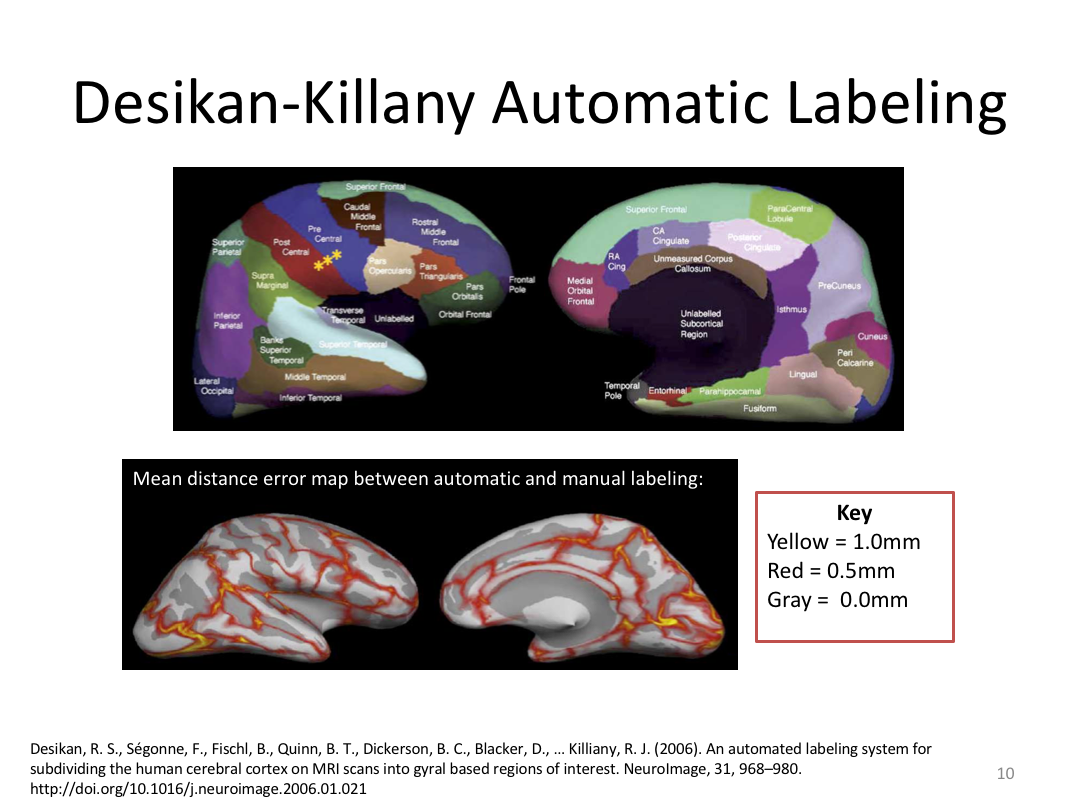
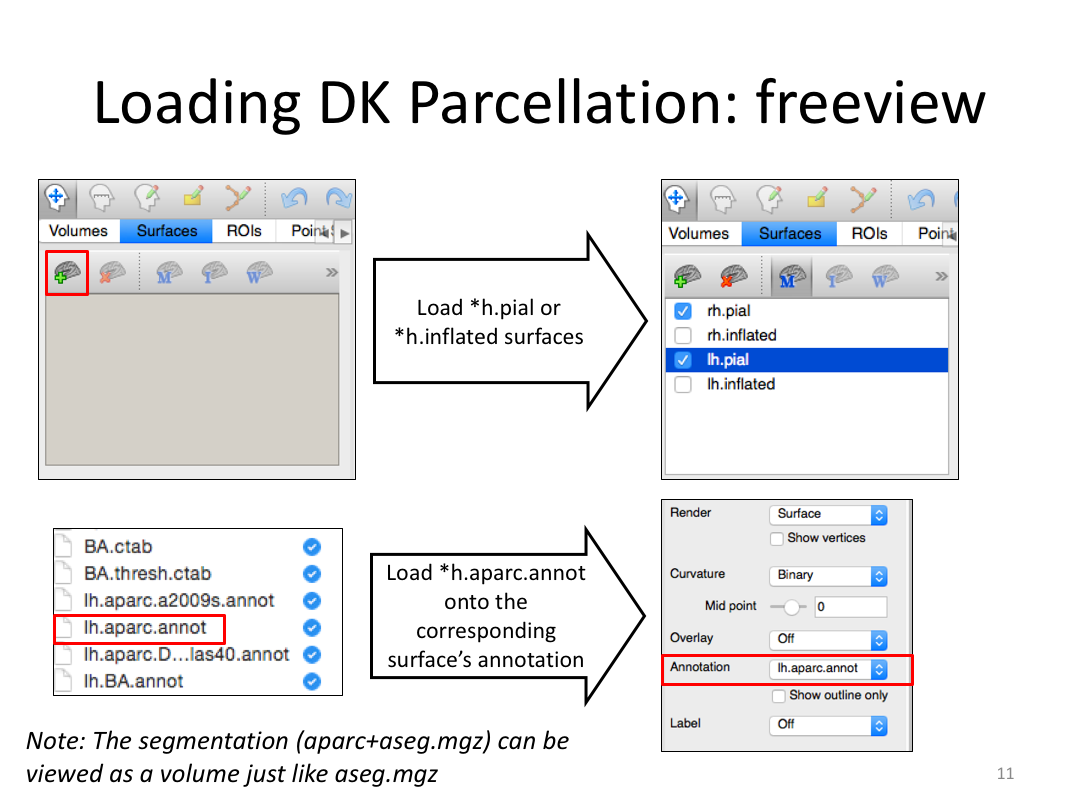


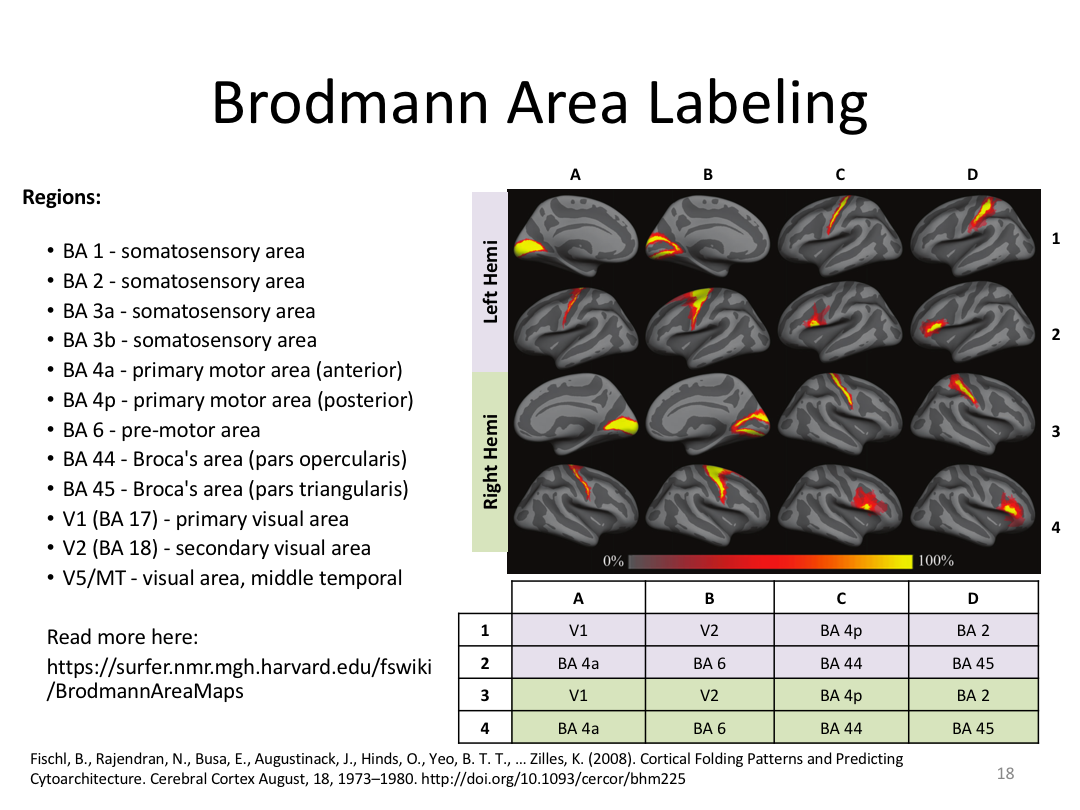
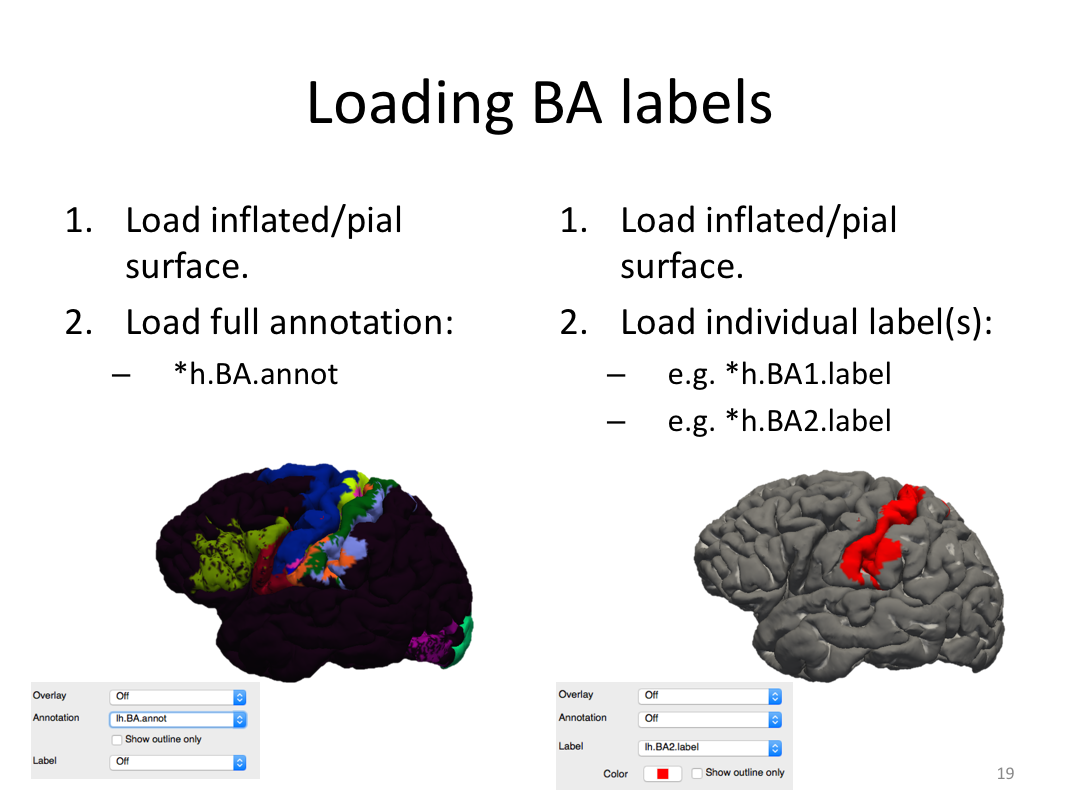
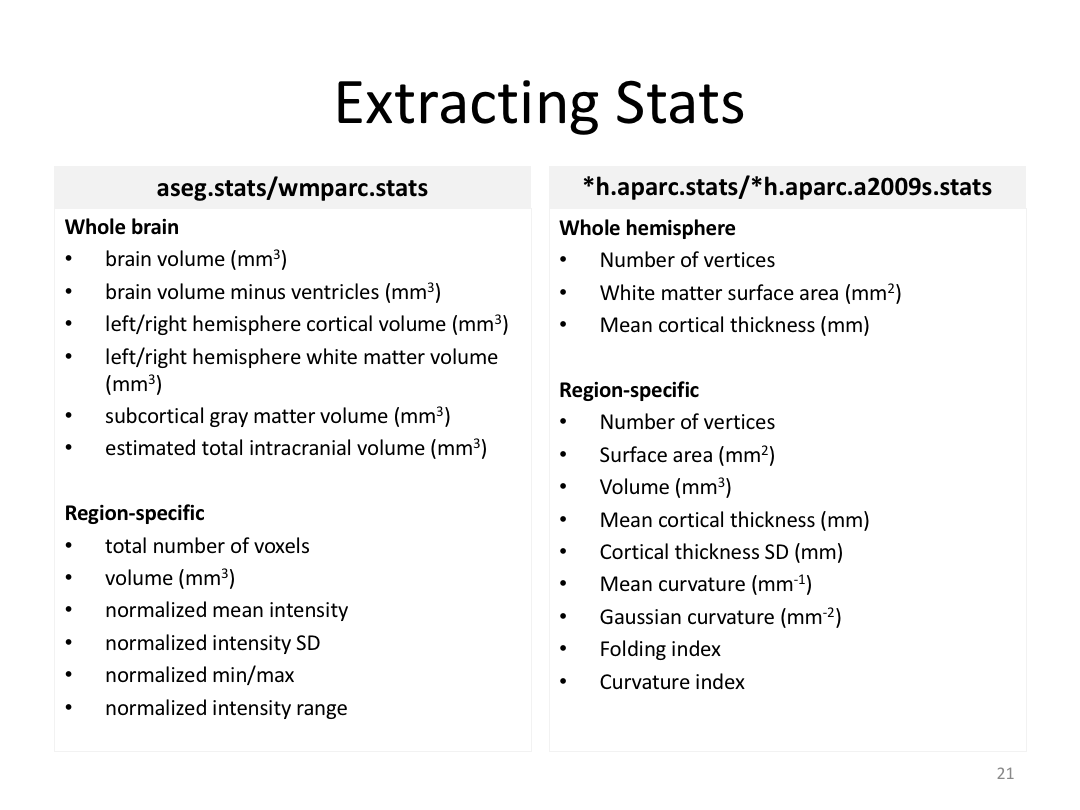


Class 4: Parcellating the Cortex (autorecon3)
Overview:
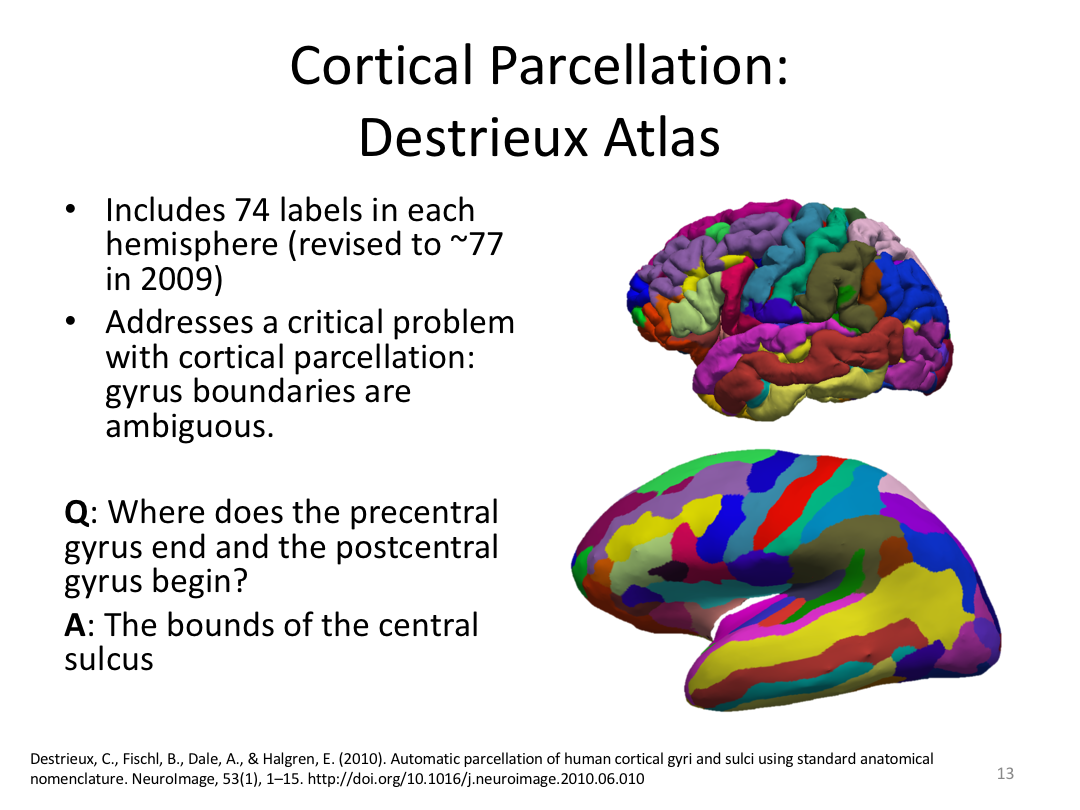
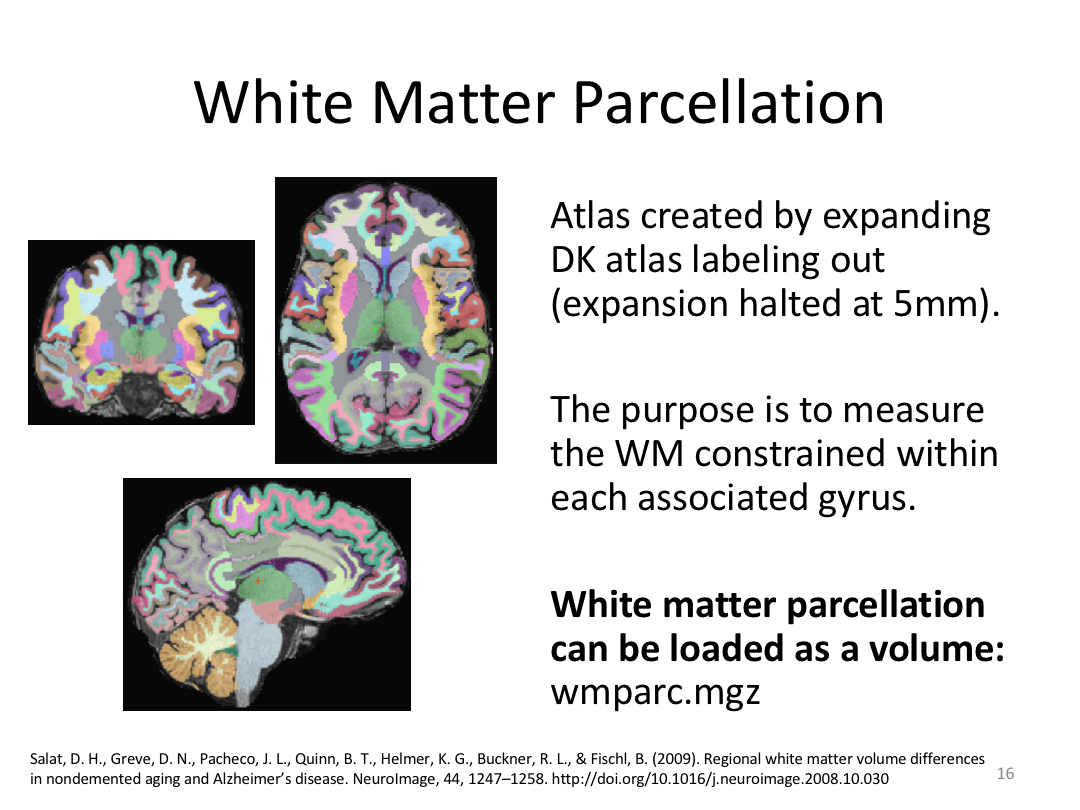
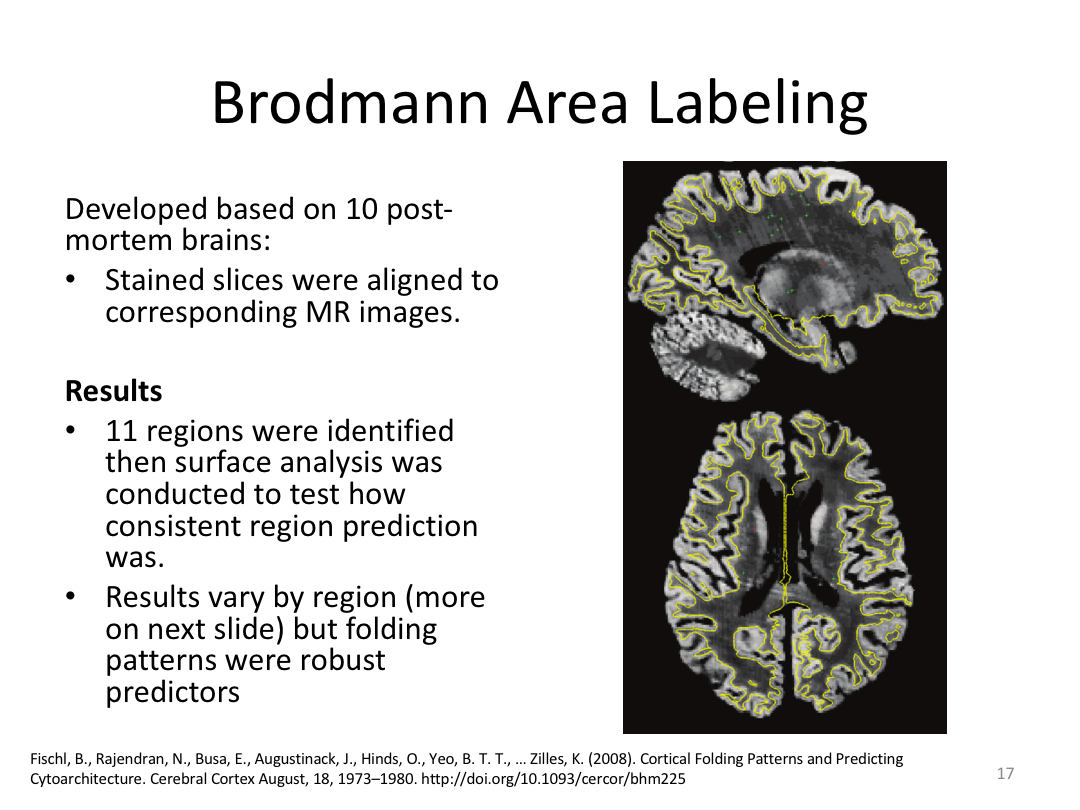
This class covered how FreeSurfer uses the surfaces created in autorecon2 to parcellate the cortex. We went over the atlases included in the software and ended with extracting data. Throughout this process, we introduced helpful features available in FreeView to visualize both the 3D cortex space and the 2D cortical segmentation.
Recommended Practice:
1. Run a brain through autorecon3
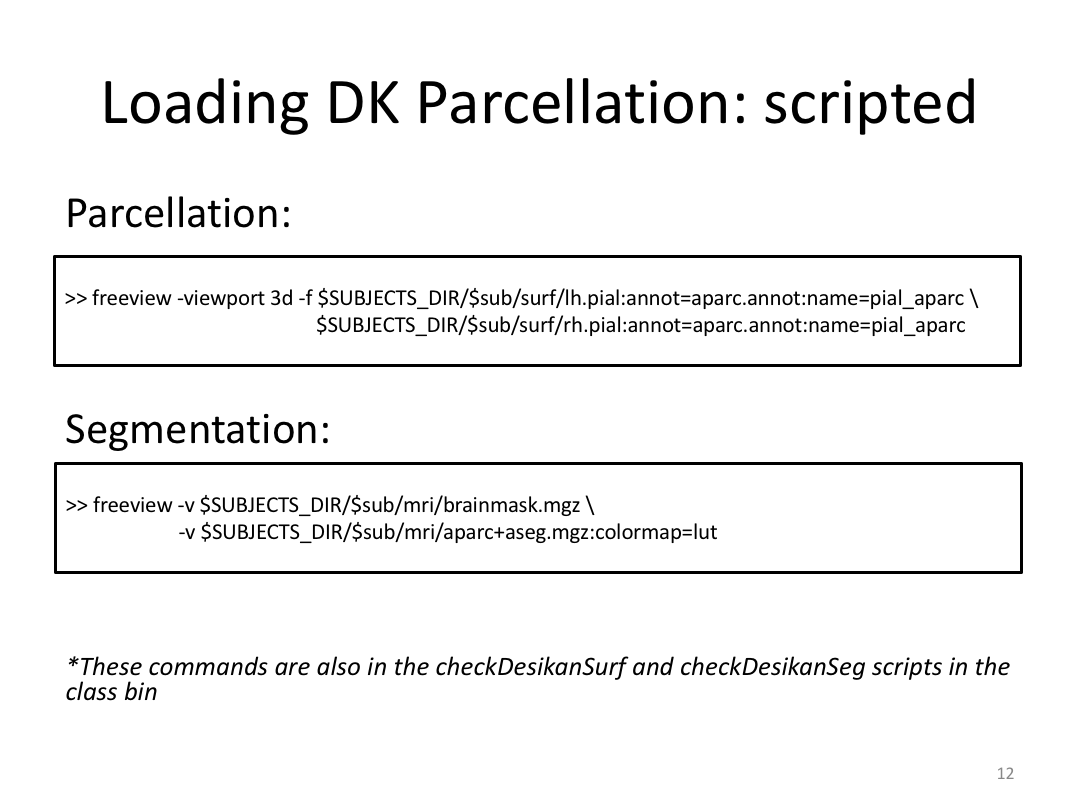
2. Open aparc+aseg.mgz and brain.mgz in freeview
3. Extract data



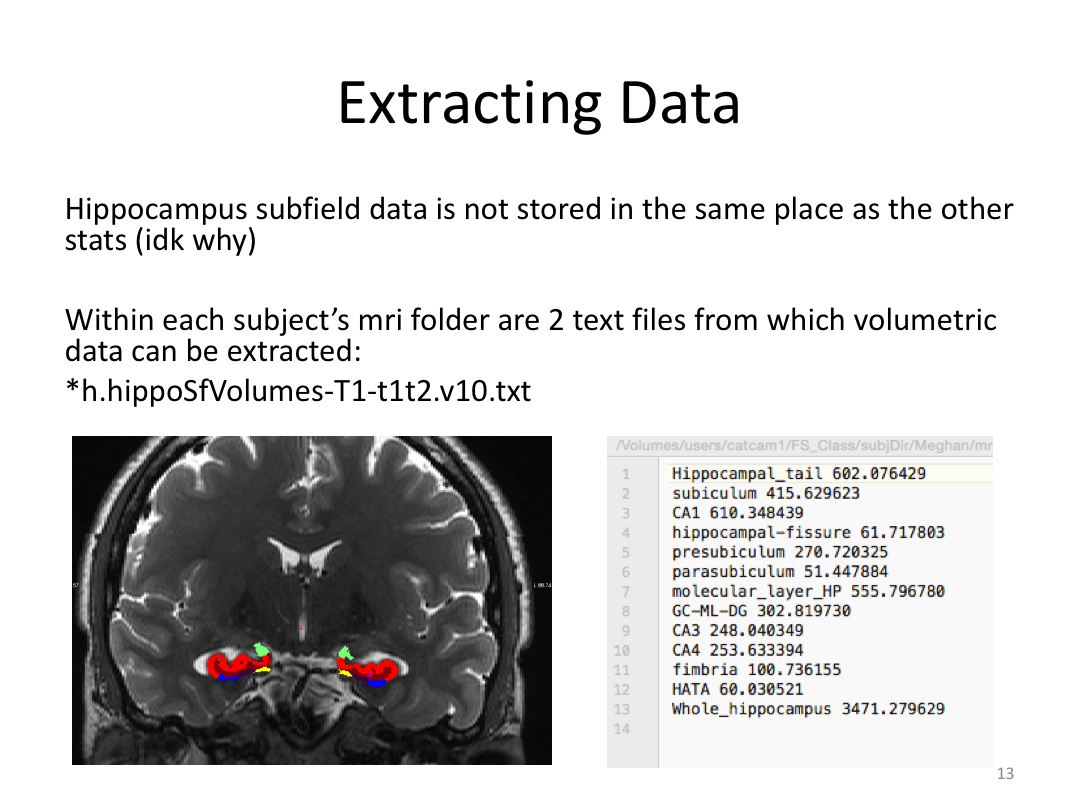

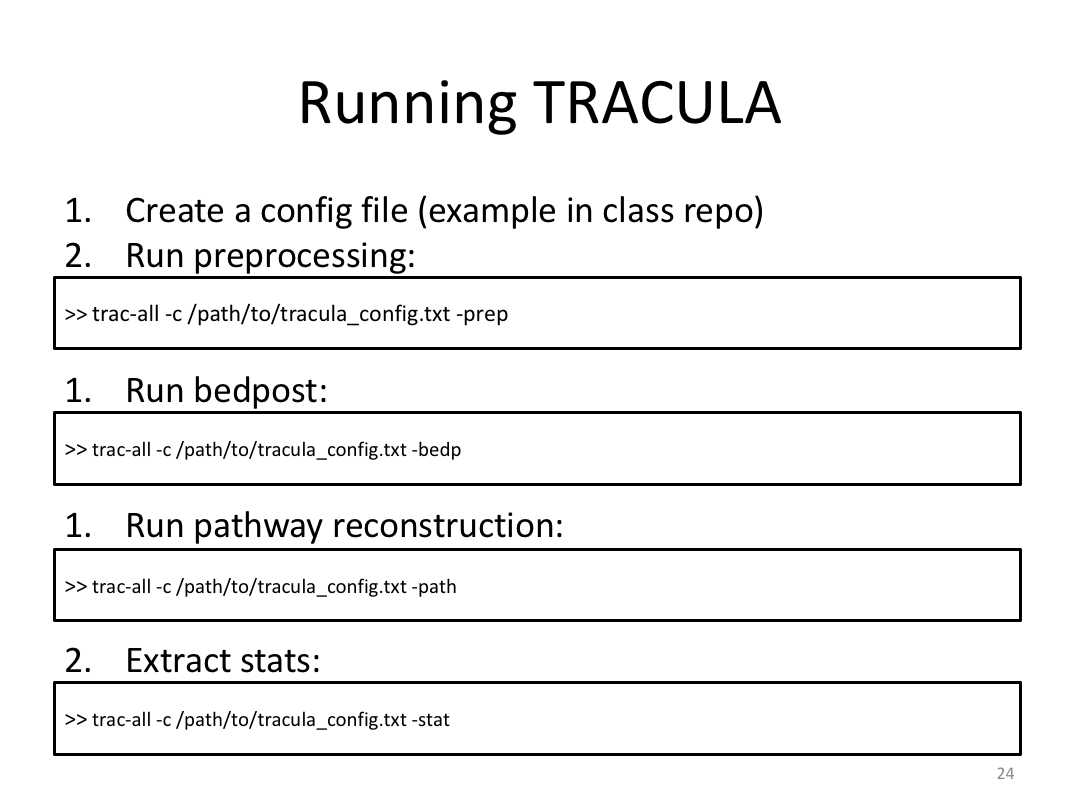



Class 5: Other Cool Stuff FreeSurfer Can Do
Overview:
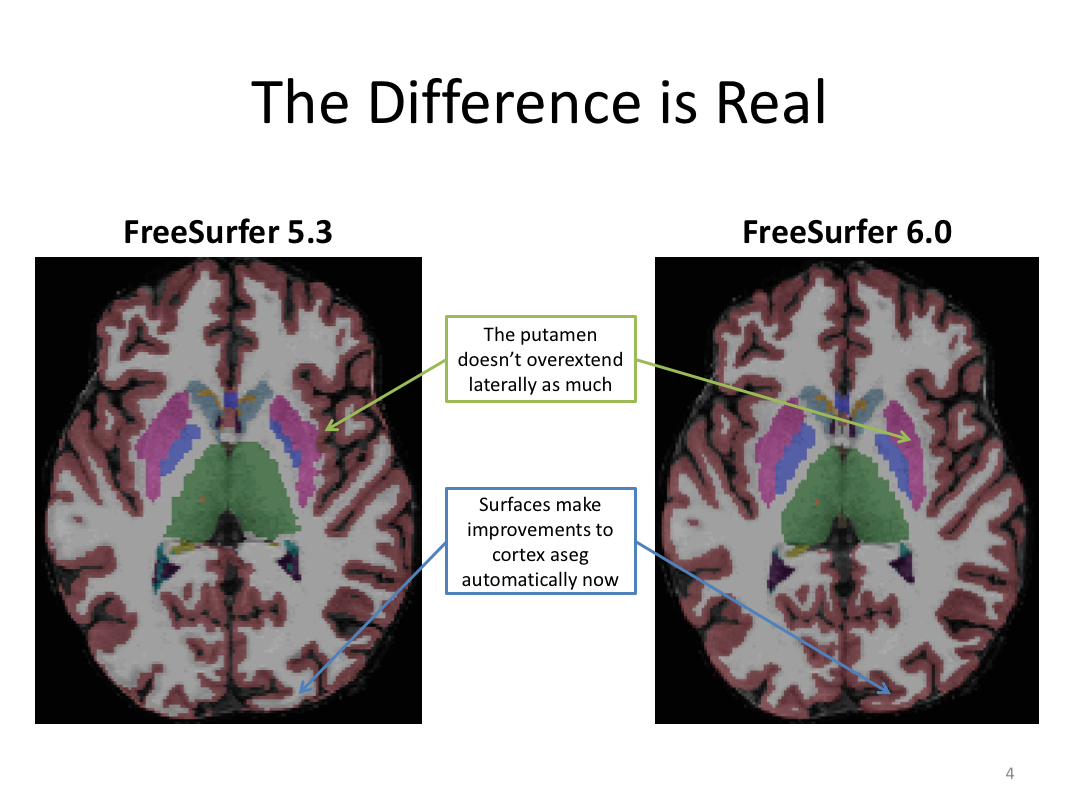
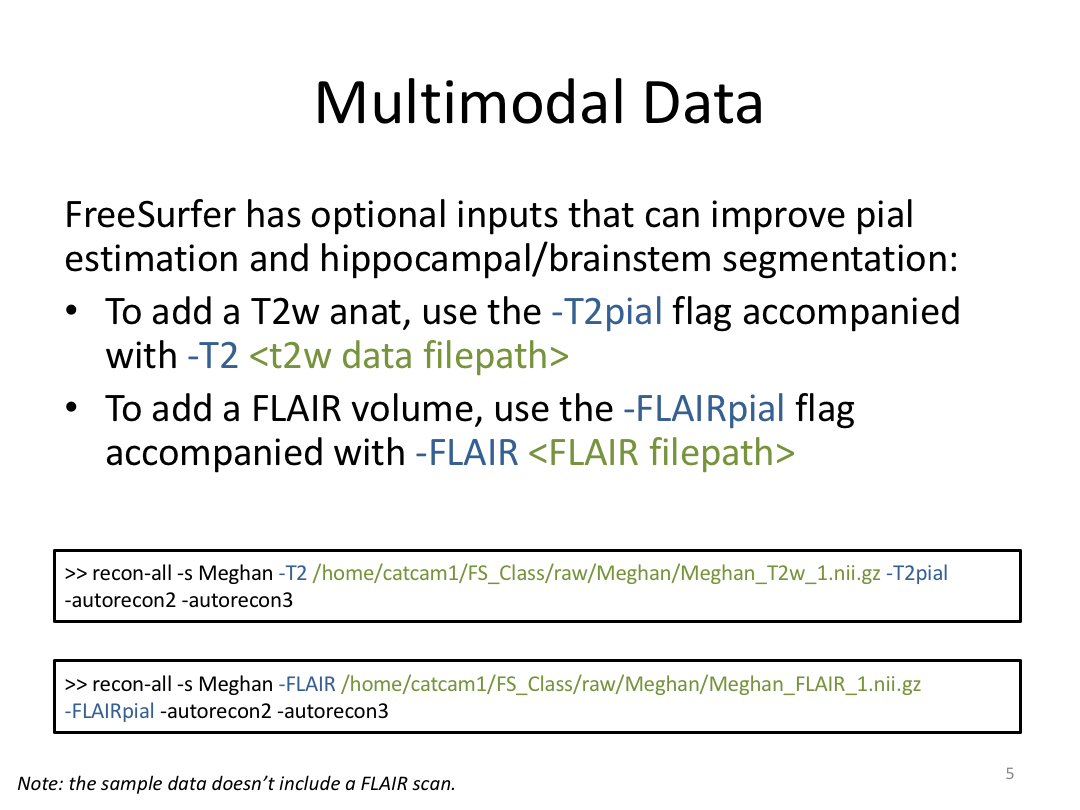
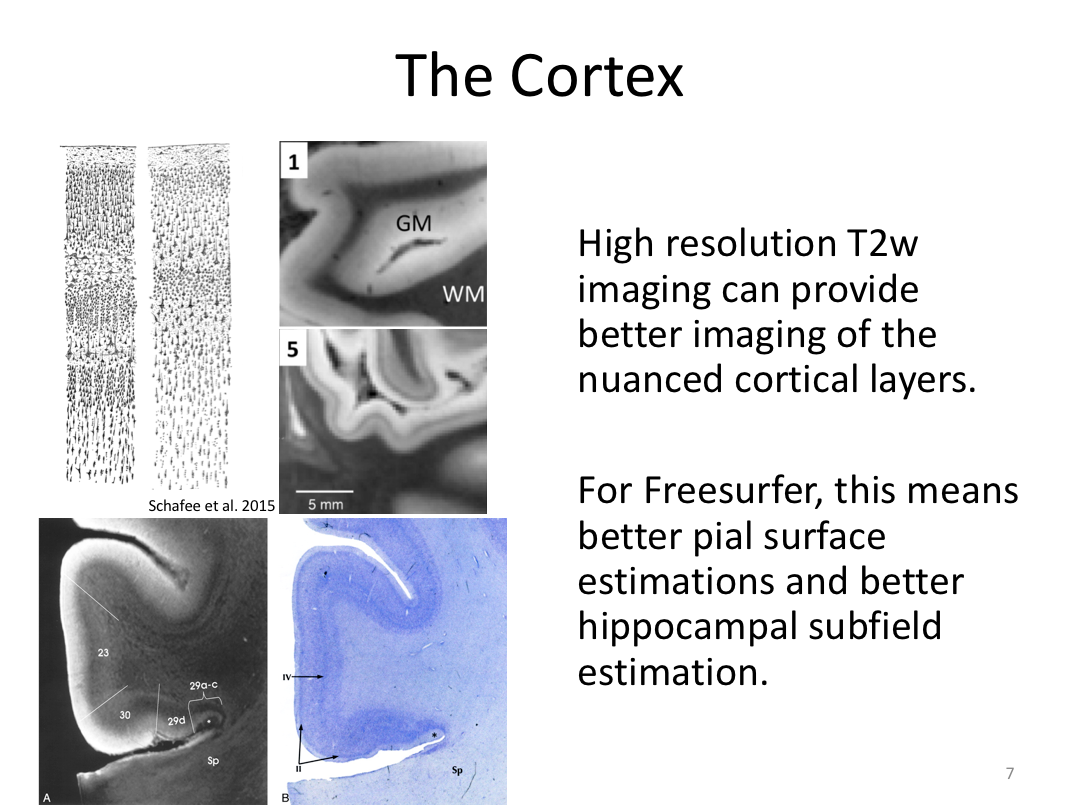
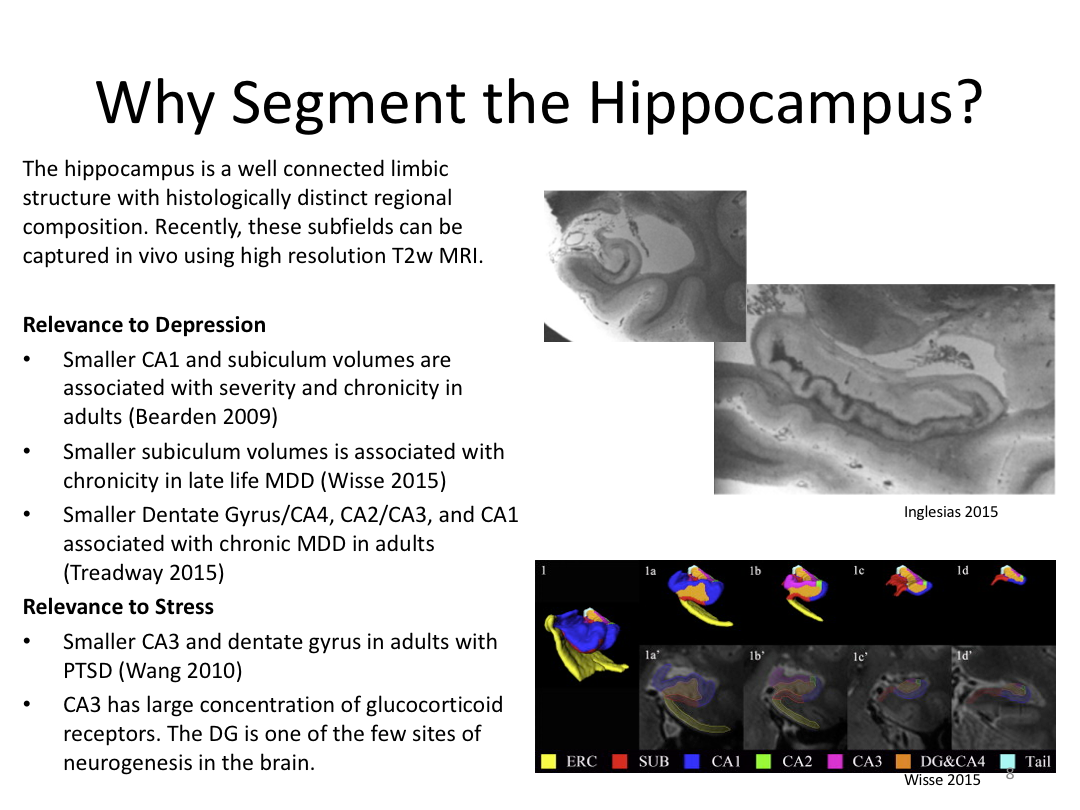
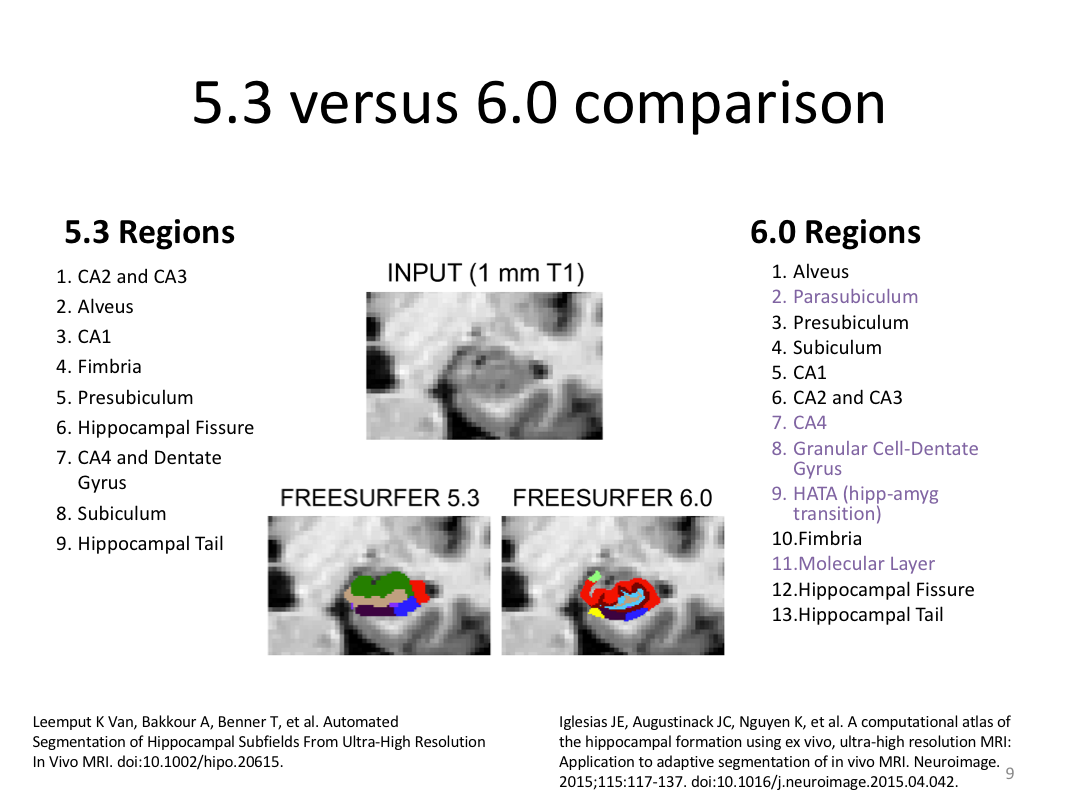
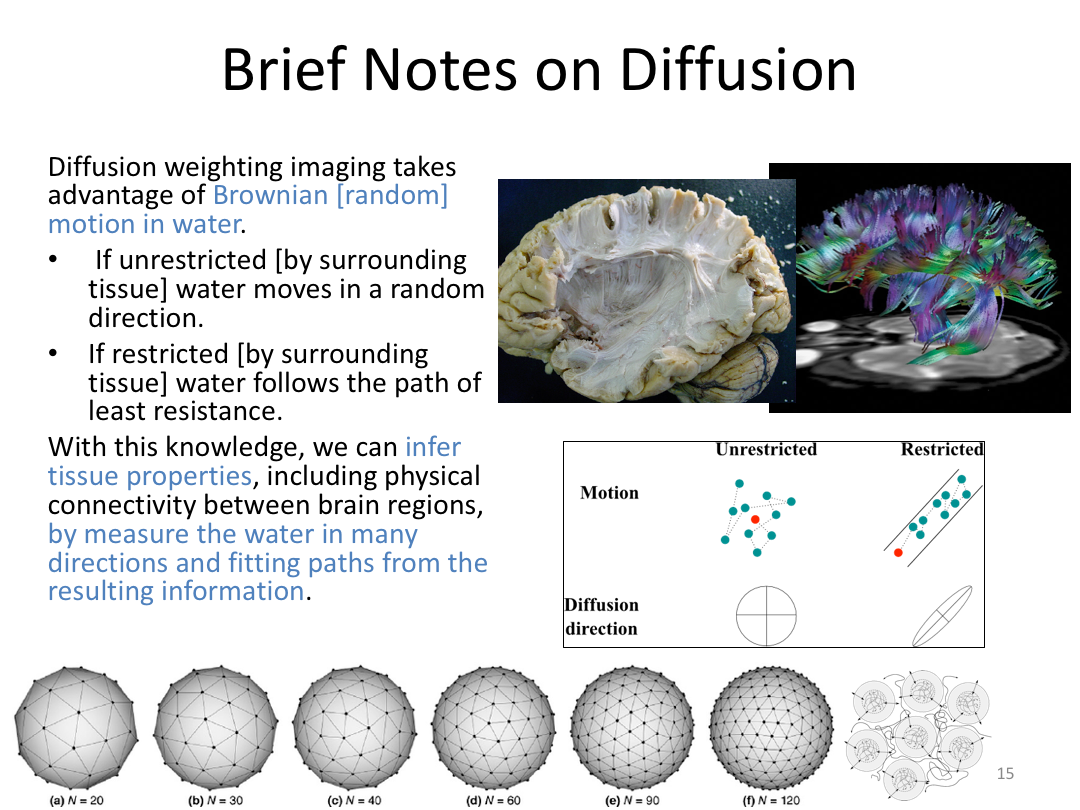
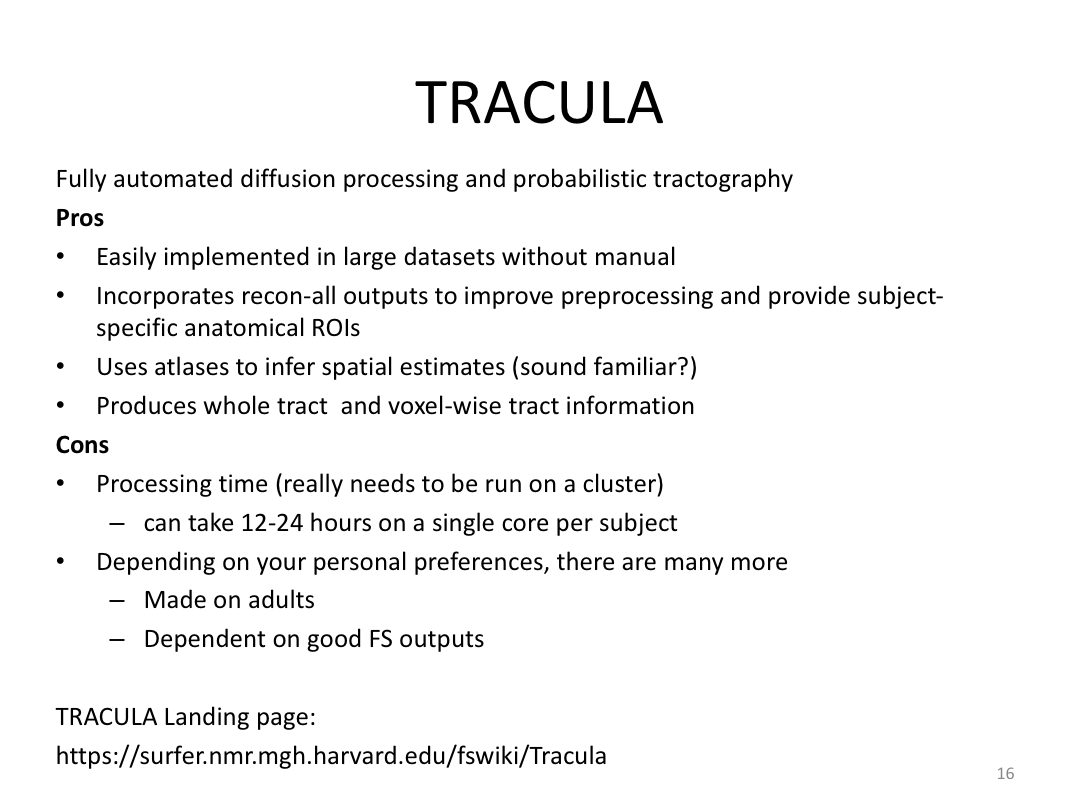
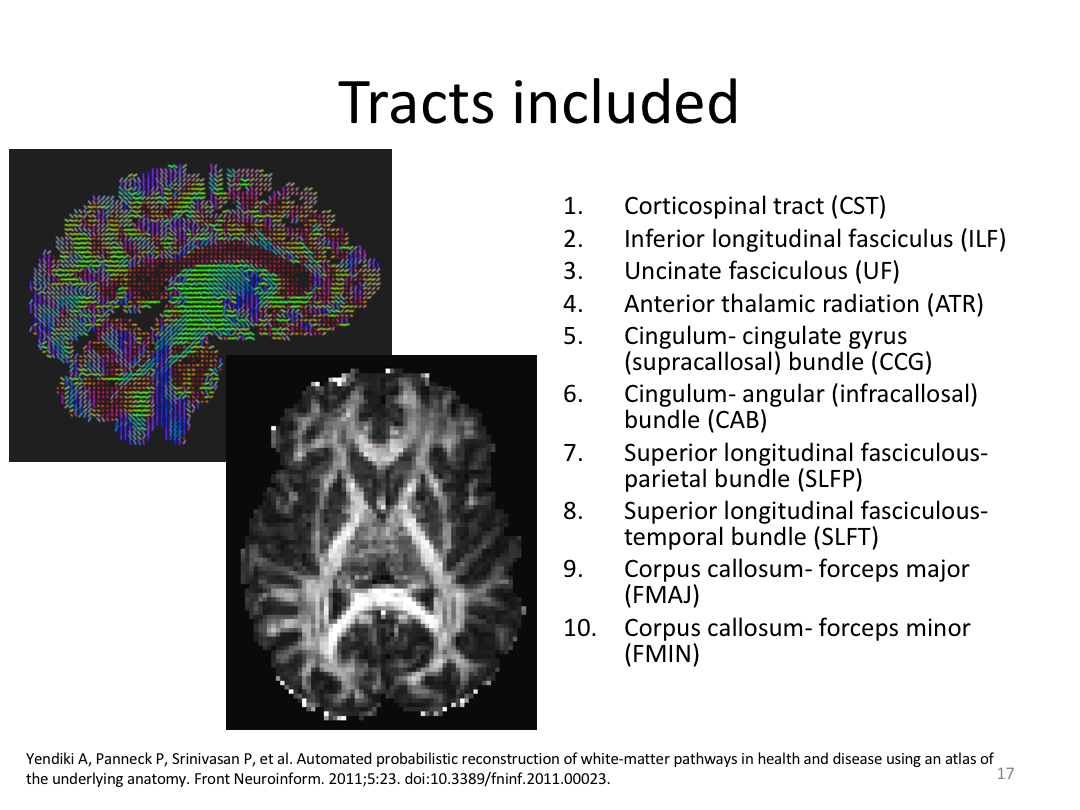
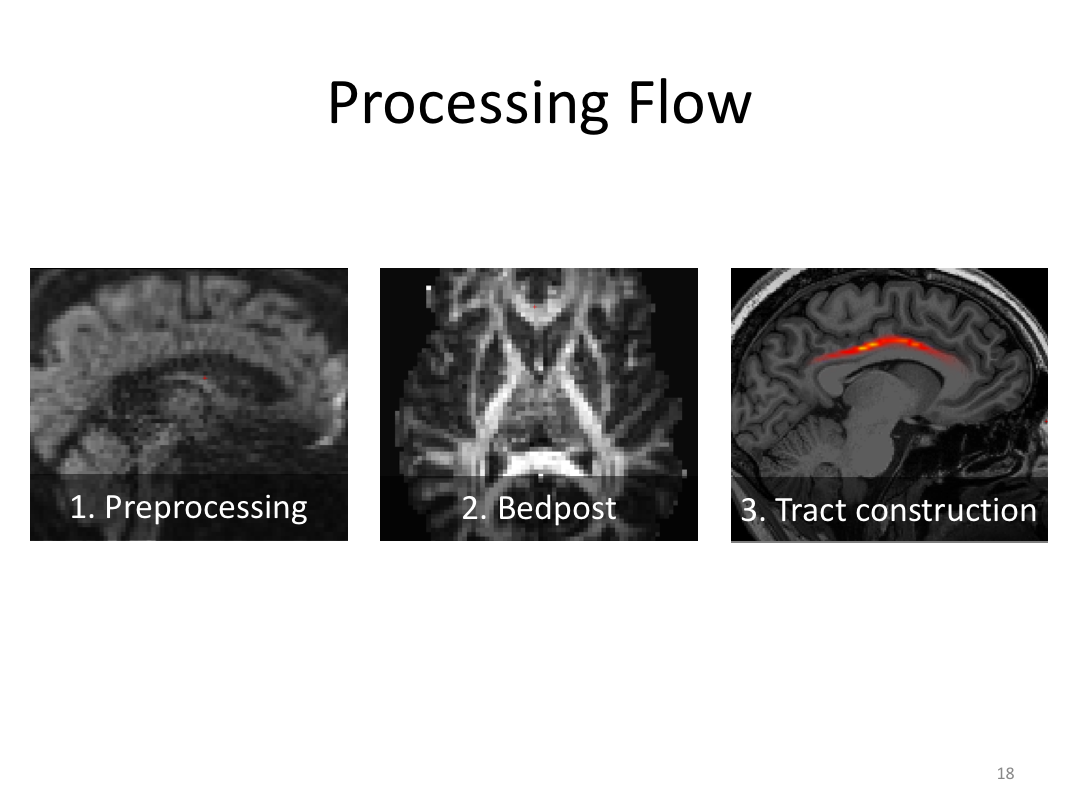
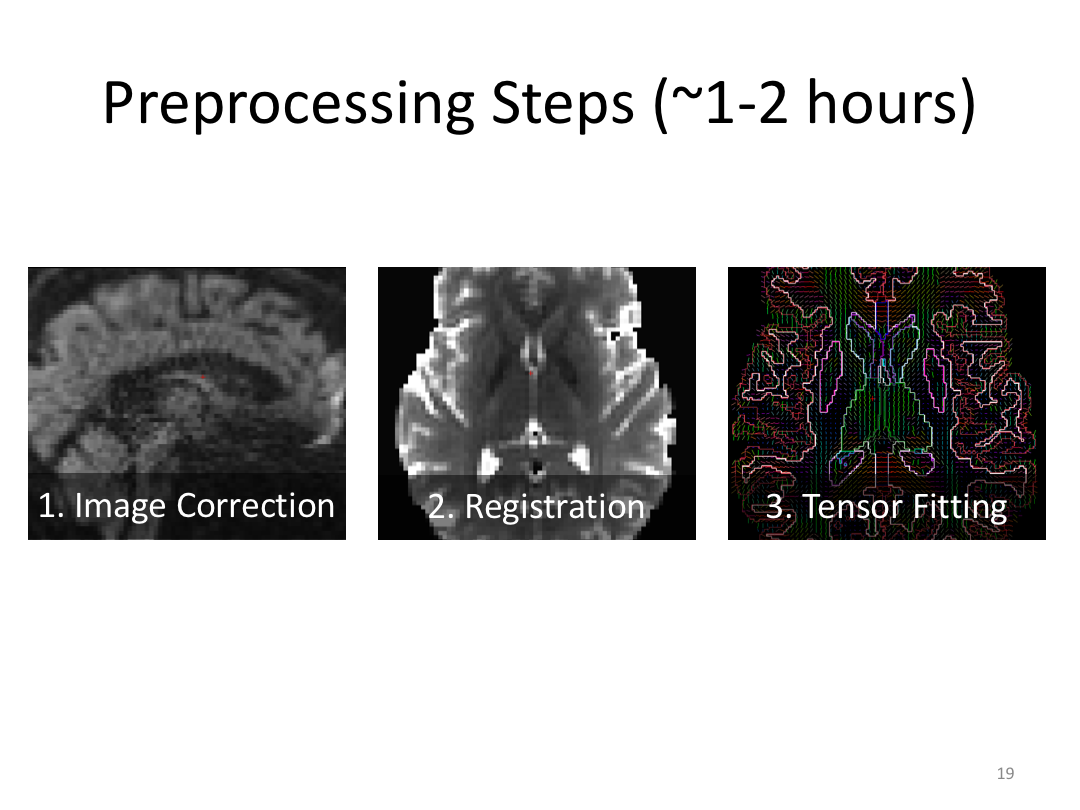
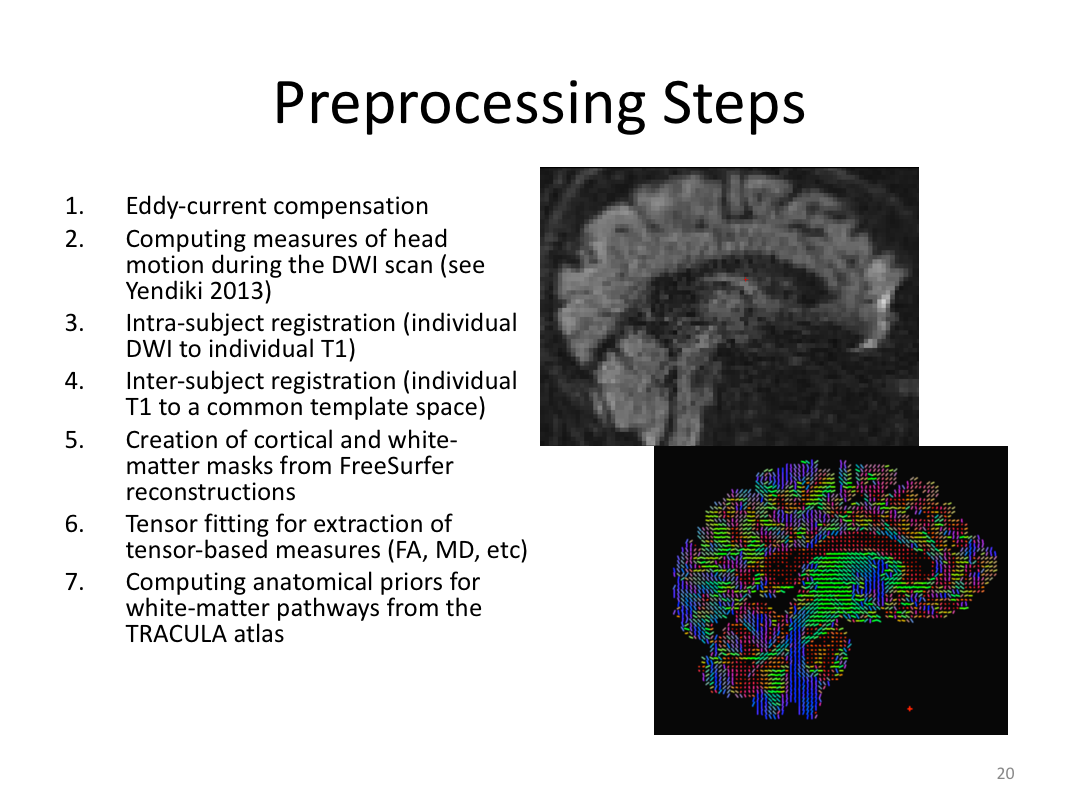
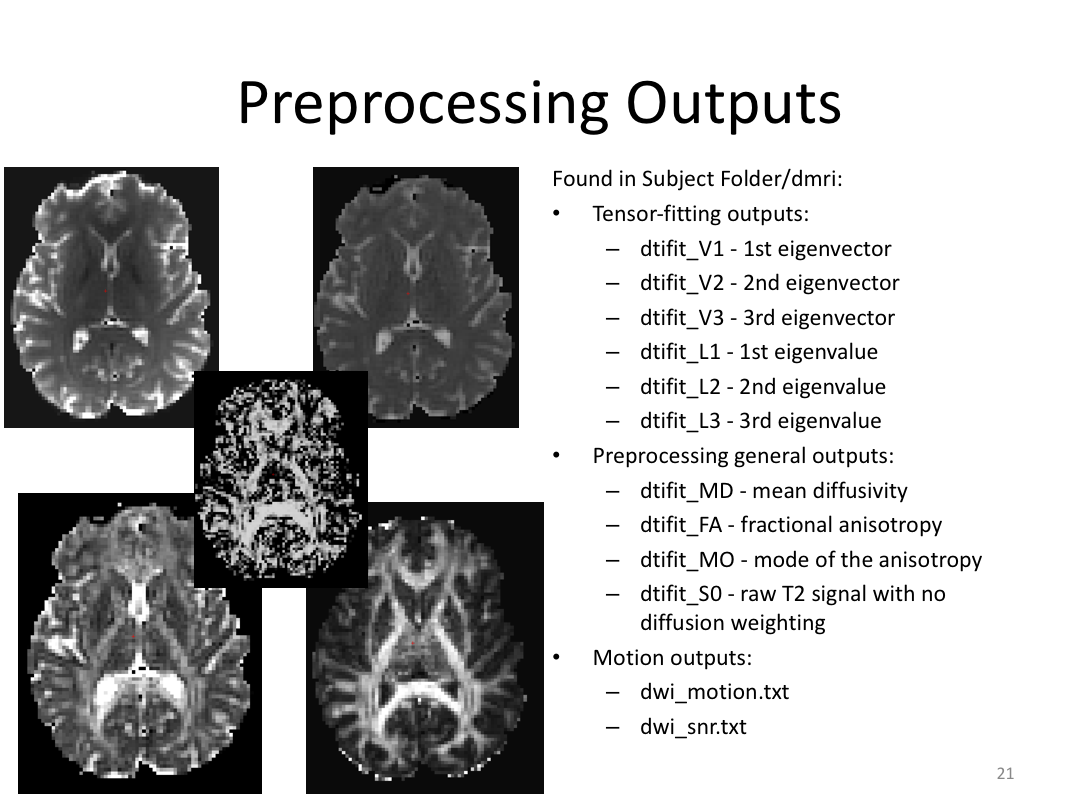
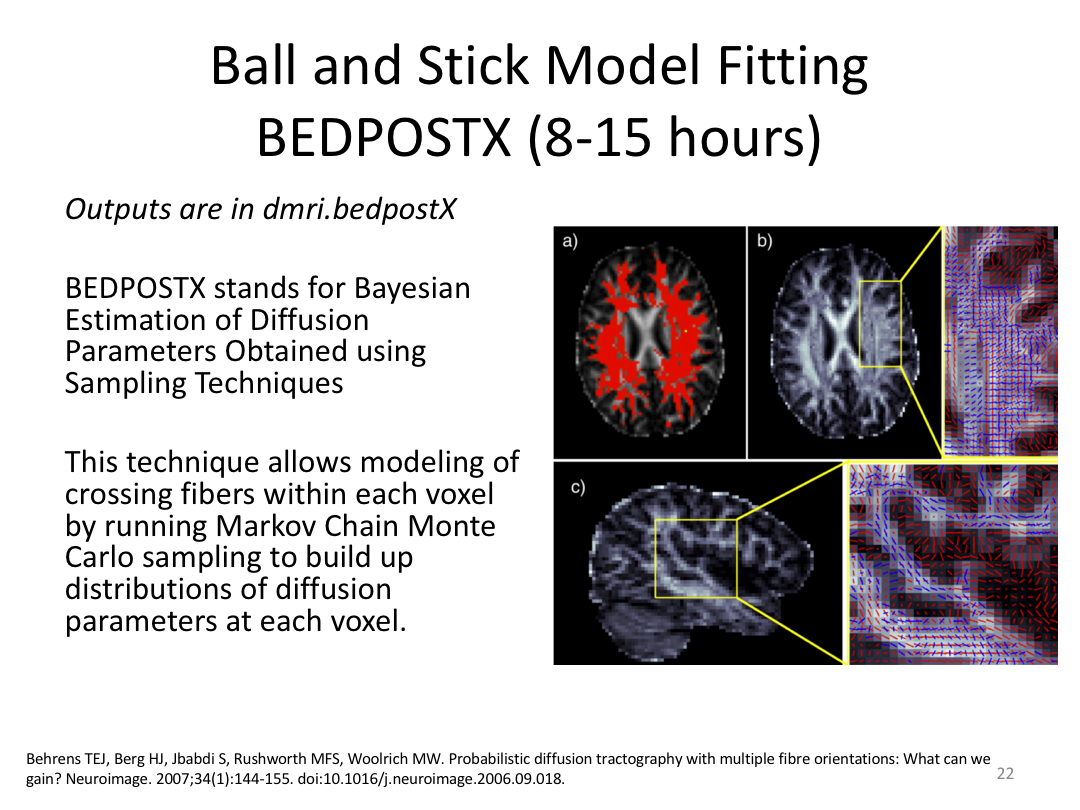
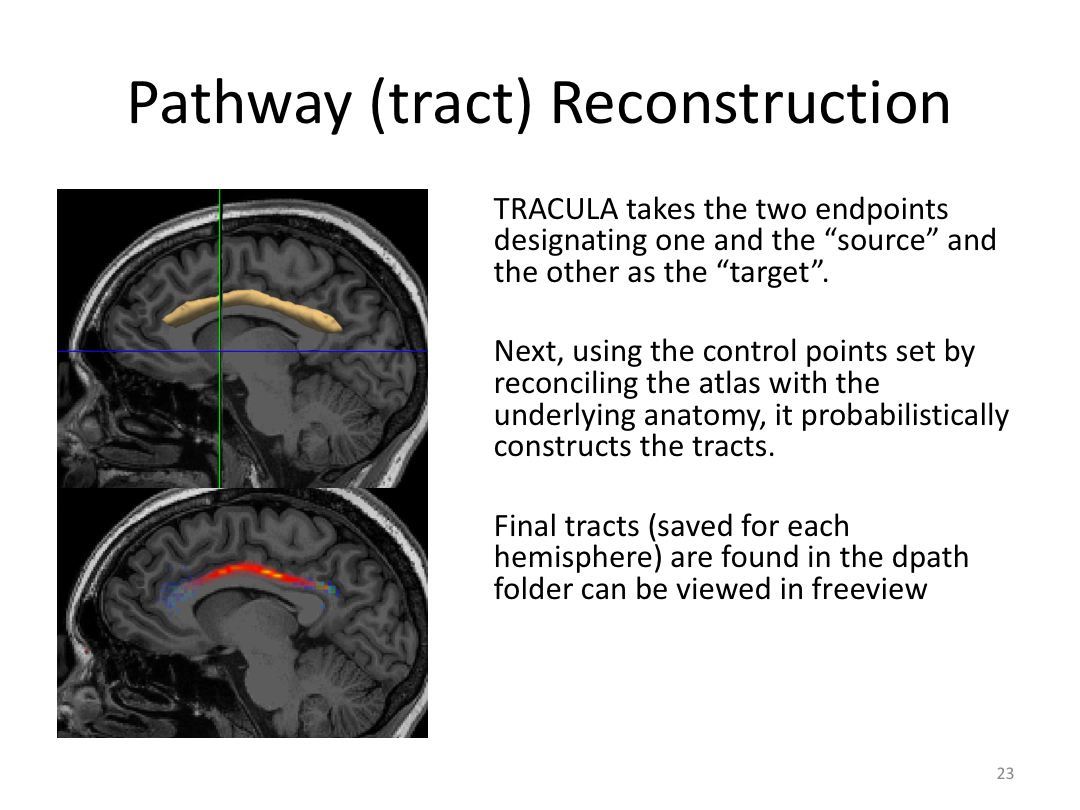
This class was optional. FreeSurfer v6.0 came out right before I taught this class, so we started with the highlights from the official release before diving into the planned class content: multimodal segmentation of the hippocampus and DTI using TRACULA.
Recommended Practice:
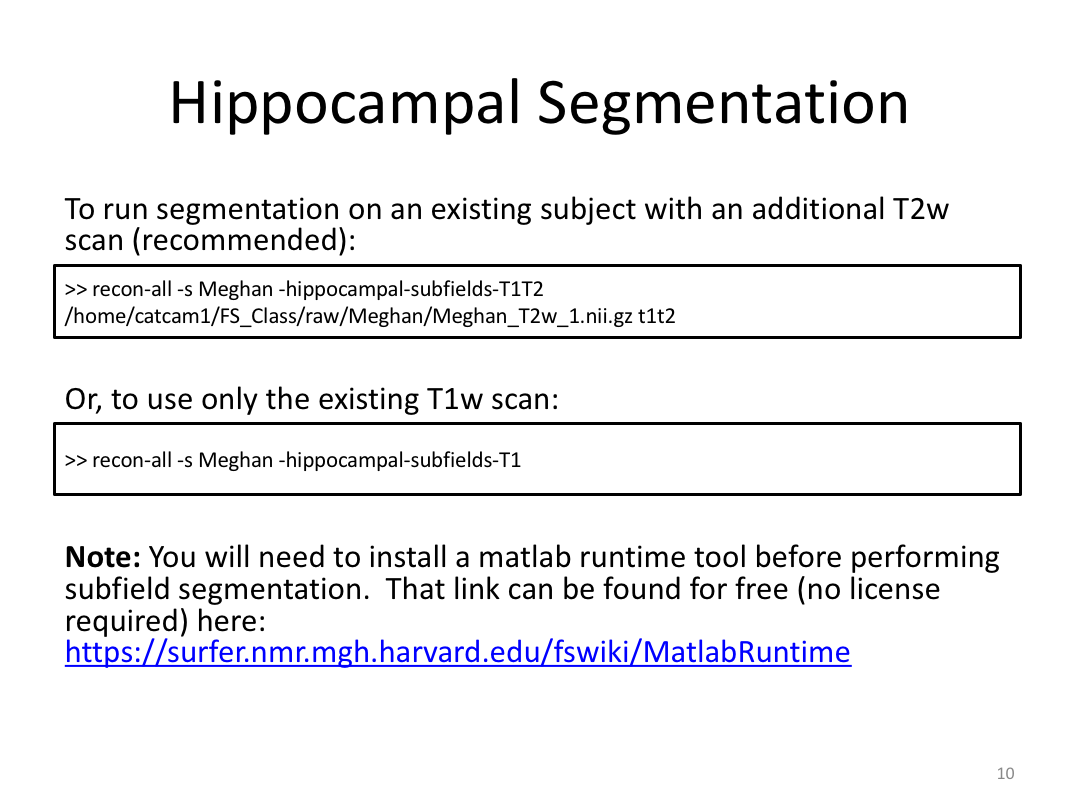
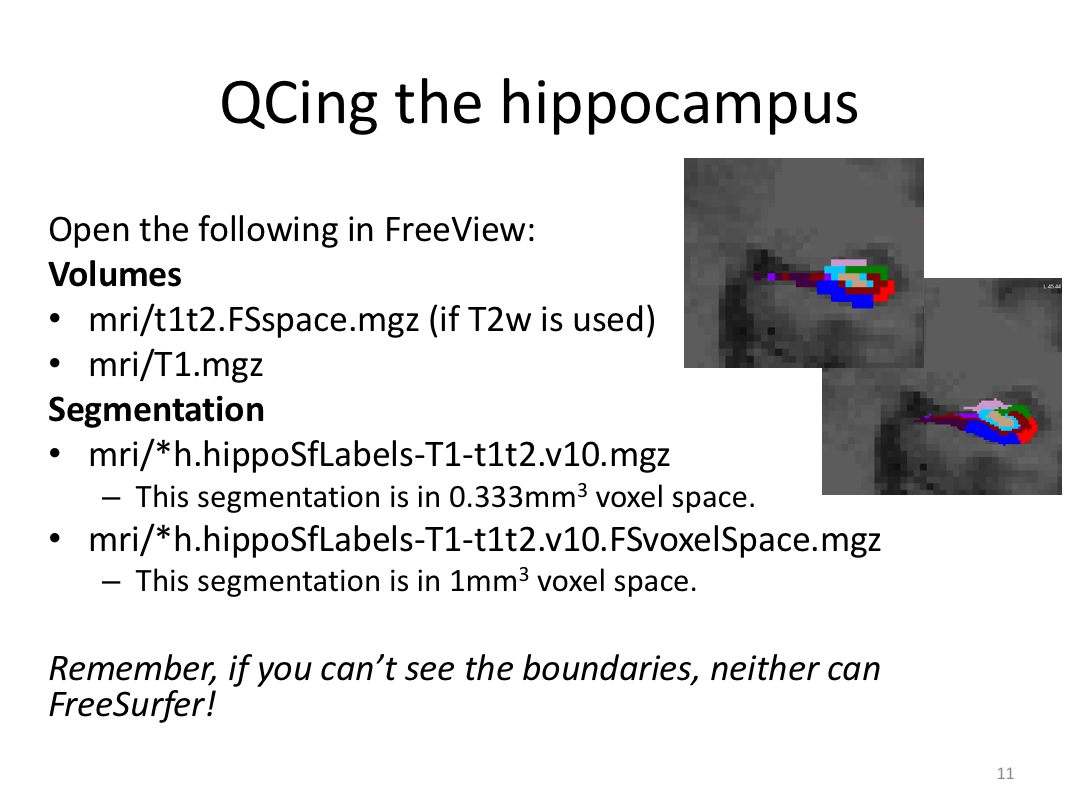
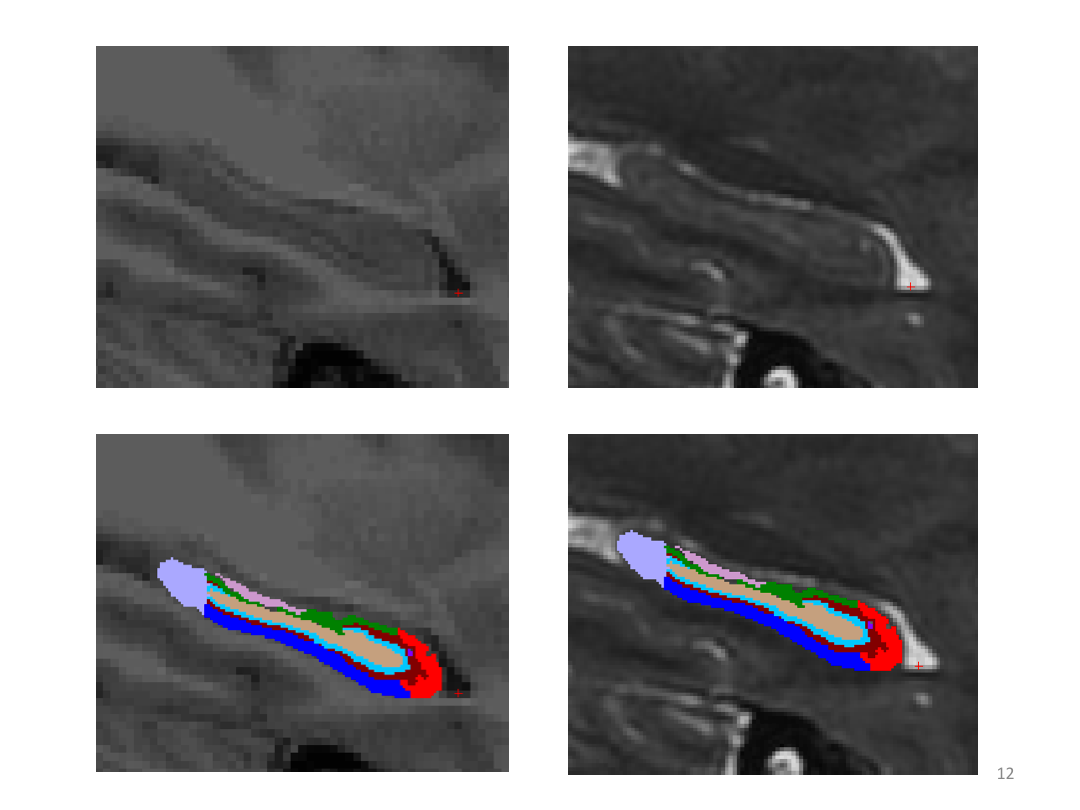
1. Segment Meghan and Monica's hippocampi (with and without a T2w image)
2. Run Meghan’s DTI brain in TRACULA
